ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1
- PMID: 25086746
- PMCID: PMC4150825
- DOI: 10.1038/ncb3013
ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1
Abstract
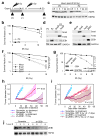
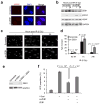
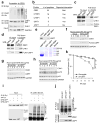
Epithelial-mesenchymal transition (EMT) is associated with characteristics of breast cancer stem cells, including chemoresistance and radioresistance. However, it is unclear whether EMT itself or specific EMT regulators play causal roles in these properties. Here we identify an EMT-inducing transcription factor, zinc finger E-box binding homeobox 1 (ZEB1), as a regulator of radiosensitivity and DNA damage response. Radioresistant subpopulations of breast cancer cells derived from ionizing radiation exhibit hyperactivation of the kinase ATM and upregulation of ZEB1, and the latter promotes tumour cell radioresistance in vitro and in vivo. Mechanistically, ATM phosphorylates and stabilizes ZEB1 in response to DNA damage, ZEB1 in turn directly interacts with USP7 and enhances its ability to deubiquitylate and stabilize CHK1, thereby promoting homologous recombination-dependent DNA repair and resistance to radiation. These findings identify ZEB1 as an ATM substrate linking ATM to CHK1 and the mechanism underlying the association between EMT and radioresistance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


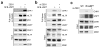

Similar articles
-
miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13.Nat Commun. 2014 Dec 5;5:5671. doi: 10.1038/ncomms6671. Nat Commun. 2014. PMID: 25476932 Free PMC article.
-
ZEB1 inhibition sensitizes cells to the ATR inhibitor VE-821 by abrogating epithelial-mesenchymal transition and enhancing DNA damage.Cell Cycle. 2018;17(5):595-604. doi: 10.1080/15384101.2017.1404206. Epub 2018 Apr 2. Cell Cycle. 2018. PMID: 29157079 Free PMC article.
-
Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors.Oncotarget. 2015 Jan 20;6(2):979-94. doi: 10.18632/oncotarget.2825. Oncotarget. 2015. PMID: 25460509 Free PMC article.
-
ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance.Cell Cycle. 2015;14(4):481-7. doi: 10.1080/15384101.2015.1006048. Cell Cycle. 2015. PMID: 25607528 Free PMC article. Review.
-
ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance.Front Mol Biosci. 2020 Mar 19;7:36. doi: 10.3389/fmolb.2020.00036. eCollection 2020. Front Mol Biosci. 2020. PMID: 32266287 Free PMC article. Review.
Cited by
-
miR-1226-5p is involved in radioresistance of colorectal cancer by activating M2 macrophages through suppressing IRF1.J Transl Med. 2024 Oct 29;22(1):980. doi: 10.1186/s12967-024-05797-1. J Transl Med. 2024. PMID: 39472937 Free PMC article.
-
Tumor Hybrid Cells: Nature and Biological Significance.Front Cell Dev Biol. 2022 Feb 15;10:814714. doi: 10.3389/fcell.2022.814714. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35242760 Free PMC article. Review.
-
RHOJ controls EMT-associated resistance to chemotherapy.Nature. 2023 Apr;616(7955):168-175. doi: 10.1038/s41586-023-05838-7. Epub 2023 Mar 22. Nature. 2023. PMID: 36949199 Free PMC article.
-
Boolean modeling of mechanosensitive epithelial to mesenchymal transition and its reversal.iScience. 2023 Mar 2;26(4):106321. doi: 10.1016/j.isci.2023.106321. eCollection 2023 Apr 21. iScience. 2023. PMID: 36968076 Free PMC article.
-
Role of Damage DNA-Binding Protein 1 in Pancreatic Cancer Progression and Chemoresistance.Cancers (Basel). 2019 Dec 12;11(12):1998. doi: 10.3390/cancers11121998. Cancers (Basel). 2019. PMID: 31842285 Free PMC article.
References
-
- Bedford JS. Sublethal damage, potentially lethal damage, and chromosomal aberrations in mammalian cells exposed to ionizing radiations. Int J Radiat Oncol Biol Phys. 1991;21:1457–1469. - PubMed
-
- Frankenberg-Schwager M, Frankenberg D, Blocher D, Adamczyk C. Effect of dose rate on the induction of DNA double-strand breaks in eucaryotic cells. Radiat Res. 1981;87:710–717. - PubMed
-
- Buchholz TA. Radiation therapy for early-stage breast cancer after breast-conserving surgery. N Engl J Med. 2009;360:63–70. - PubMed
-
- Jameel JK, Rao VS, Cawkwell L, Drew PJ. Radioresistance in carcinoma of the breast. Breast. 2004;13:452–460. - PubMed
-
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

