A framework for the interpretation of de novo mutation in human disease
- PMID: 25086666
- PMCID: PMC4222185
- DOI: 10.1038/ng.3050
A framework for the interpretation of de novo mutation in human disease
Abstract
Spontaneously arising (de novo) mutations have an important role in medical genetics. For diseases with extensive locus heterogeneity, such as autism spectrum disorders (ASDs), the signal from de novo mutations is distributed across many genes, making it difficult to distinguish disease-relevant mutations from background variation. Here we provide a statistical framework for the analysis of excesses in de novo mutation per gene and gene set by calibrating a model of de novo mutation. We applied this framework to de novo mutations collected from 1,078 ASD family trios, and, whereas we affirmed a significant role for loss-of-function mutations, we found no excess of de novo loss-of-function mutations in cases with IQ above 100, suggesting that the role of de novo mutations in ASDs might reside in fundamental neurodevelopmental processes. We also used our model to identify ∼1,000 genes that are significantly lacking in functional coding variation in non-ASD samples and are enriched for de novo loss-of-function mutations identified in ASD cases.
Conflict of interest statement
Figures
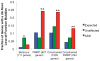
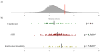
Similar articles
-
Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations.Nat Genet. 2011 Jun;43(6):585-9. doi: 10.1038/ng.835. Epub 2011 May 15. Nat Genet. 2011. PMID: 21572417 Free PMC article.
-
Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes.PLoS Genet. 2013;9(8):e1003671. doi: 10.1371/journal.pgen.1003671. Epub 2013 Aug 15. PLoS Genet. 2013. PMID: 23966865 Free PMC article.
-
Autism spectrum disorder severity reflects the average contribution of de novo and familial influences.Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):15161-5. doi: 10.1073/pnas.1409204111. Epub 2014 Oct 6. Proc Natl Acad Sci U S A. 2014. PMID: 25288738 Free PMC article.
-
The role of de novo mutations in the genetics of autism spectrum disorders.Nat Rev Genet. 2014 Feb;15(2):133-41. doi: 10.1038/nrg3585. Epub 2014 Jan 16. Nat Rev Genet. 2014. PMID: 24430941 Review.
-
From de novo mutations to personalized therapeutic interventions in autism.Annu Rev Med. 2015;66:487-507. doi: 10.1146/annurev-med-091113-024550. Annu Rev Med. 2015. PMID: 25587659 Review.
Cited by
-
Compendium of proteins containing segments that exhibit zero-tolerance to amino acid variation in humans.Protein Sci. 2022 Sep;31(9):e4408. doi: 10.1002/pro.4408. Protein Sci. 2022. PMID: 36040257 Free PMC article.
-
Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability.Nat Neurosci. 2016 Sep;19(9):1194-6. doi: 10.1038/nn.4352. Epub 2016 Aug 1. Nat Neurosci. 2016. PMID: 27479843 Review.
-
De novo missense variants in HECW2 are associated with neurodevelopmental delay and hypotonia.J Med Genet. 2017 Feb;54(2):84-86. doi: 10.1136/jmedgenet-2016-103943. Epub 2016 Jul 7. J Med Genet. 2017. PMID: 27389779 Free PMC article.
-
Long-read genome sequencing for the molecular diagnosis of neurodevelopmental disorders.HGG Adv. 2021 Apr 8;2(2):100023. doi: 10.1016/j.xhgg.2021.100023. Epub 2021 Jan 16. HGG Adv. 2021. PMID: 33937879 Free PMC article.
-
Focused Strategies for Defining the Genetic Architecture of Congenital Heart Defects.Genes (Basel). 2021 May 28;12(6):827. doi: 10.3390/genes12060827. Genes (Basel). 2021. PMID: 34071175 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
- U24 MH081810/MH/NIMH NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- R01 MH089004/MH/NIMH NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- R01 MH057881/MH/NIMH NIH HHS/United States
- HL-102924/HL/NHLBI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- U24 MH068457/MH/NIMH NIH HHS/United States
- HL-102925/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- R01 MH089482/MH/NIMH NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102926/HL/NHLBI NIH HHS/United States
- R01 MH089208/MH/NIMH NIH HHS/United States
- UC2 HL103010/HL/NHLBI NIH HHS/United States
- HL-103010/HL/NHLBI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- U01 MH100229/MH/NIMH NIH HHS/United States
- P50HD055751/HD/NICHD NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- R01 MH061009/MH/NIMH NIH HHS/United States
- U24MH068457/MH/NIMH NIH HHS/United States
- 1U24MH081810/MH/NIMH NIH HHS/United States
- R01MH061009/MH/NIMH NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01MH089175/MH/NIMH NIH HHS/United States
- P50 HD055751/HD/NICHD NIH HHS/United States
- HL-102926/HL/NHLBI NIH HHS/United States
- R01 MH089025/MH/NIMH NIH HHS/United States
- R01MH089004/MH/NIMH NIH HHS/United States
- R01MH089208/MH/NIMH NIH HHS/United States
- R01 MH089175/MH/NIMH NIH HHS/United States
- R01MH057881/MH/NIMH NIH HHS/United States
- UC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102924/HL/NHLBI NIH HHS/United States
- HL-102923/HL/NHLBI NIH HHS/United States
- R01MH089025/MH/NIMH NIH HHS/United States
- R01MH089482/MH/NIMH NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UC2 HL102925/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

