Structural and functional coordination of DNA and histone methylation
- PMID: 25085914
- PMCID: PMC4107986
- DOI: 10.1101/cshperspect.a018747
Structural and functional coordination of DNA and histone methylation
Abstract
One of the most fundamental questions in the control of gene expression in mammals is how epigenetic methylation patterns of DNA and histones are established, erased, and recognized. This central process in controlling gene expression includes coordinated covalent modifications of DNA and its associated histones. This article focuses on structural aspects of enzymatic activities of histone (arginine and lysine) methylation and demethylation and functional links between the methylation status of the DNA and histones. An interconnected network of methyltransferases, demethylases, and accessory proteins is responsible for changing or maintaining the modification status of specific regions of chromatin.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
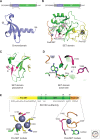



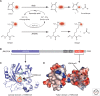




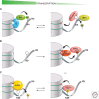
Similar articles
-
Coordinated chromatin control: structural and functional linkage of DNA and histone methylation.Biochemistry. 2010 Apr 13;49(14):2999-3008. doi: 10.1021/bi100213t. Biochemistry. 2010. PMID: 20210320 Free PMC article. Review.
-
Dynamic protein methylation in chromatin biology.Cell Mol Life Sci. 2009 Feb;66(3):407-22. doi: 10.1007/s00018-008-8303-z. Cell Mol Life Sci. 2009. PMID: 18923809 Free PMC article. Review.
-
Cross-talk among epigenetic modifications: lessons from histone arginine methylation.Biochem Soc Trans. 2013 Jun;41(3):751-9. doi: 10.1042/BST20130003. Biochem Soc Trans. 2013. PMID: 23697934 Review.
-
The control of histone lysine methylation in epigenetic regulation.Biochimie. 2007 Jan;89(1):1-20. doi: 10.1016/j.biochi.2006.07.009. Epub 2006 Aug 4. Biochimie. 2007. PMID: 16919862 Review.
-
The Mechanisms of Generation, Recognition, and Erasure of DNA 5-Methylcytosine and Thymine Oxidations.J Biol Chem. 2015 Aug 21;290(34):20723-20733. doi: 10.1074/jbc.R115.656884. Epub 2015 Jul 7. J Biol Chem. 2015. PMID: 26152719 Free PMC article. Review.
Cited by
-
A cross-talk between DNA methylation and H3 lysine 9 dimethylation at the KvDMR1 region controls the induction of Cdkn1c in muscle cells.Epigenetics. 2016 Nov;11(11):791-803. doi: 10.1080/15592294.2016.1230576. Epub 2016 Sep 9. Epigenetics. 2016. PMID: 27611768 Free PMC article.
-
Neuronal Lhx1 expression is regulated by DNMT1-dependent modulation of histone marks.Epigenetics. 2020 Nov;15(11):1259-1274. doi: 10.1080/15592294.2020.1767372. Epub 2020 May 22. Epigenetics. 2020. PMID: 32441560 Free PMC article.
-
Histone Modifications and Cancer.Cold Spring Harb Perspect Biol. 2016 Apr 1;8(4):a019521. doi: 10.1101/cshperspect.a019521. Cold Spring Harb Perspect Biol. 2016. PMID: 27037415 Free PMC article. Review.
-
Epigenetics. Restricted epigenetic inheritance of H3K9 methylation.Science. 2015 Apr 3;348(6230):132-5. doi: 10.1126/science.1260638. Science. 2015. PMID: 25838386 Free PMC article.
-
Identification of epigenetic dysregulation gene markers and immune landscape in kidney renal clear cell carcinoma by comprehensive genomic analysis.Front Immunol. 2022 Aug 18;13:901662. doi: 10.3389/fimmu.2022.901662. eCollection 2022. Front Immunol. 2022. PMID: 36059531 Free PMC article.
References
-
- Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al. 2008. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 27: 2187–2197 - PubMed
-
- Allis CD, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739 - DOI
-
- An W, Kim J, Roeder RG 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748 - PubMed
-
- Ansari KI, Mandal SS 2010. Mixed lineage leukemia: Roles in gene expression, hormone signaling and mRNA processing. FEBS J 277: 1790–1804 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
