Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription
- PMID: 25085421
- PMCID: PMC4117944
- DOI: 10.1101/gad.244434.114
Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription
Erratum in
- Genes Dev. 2014 Oct 1;28(19):2188
Abstract
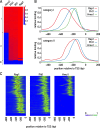
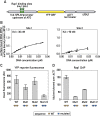
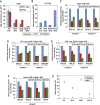
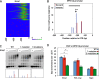
In yeast, ribosome production is controlled transcriptionally by tight coregulation of the 138 ribosomal protein genes (RPGs). RPG promoters display limited sequence homology, and the molecular basis for their coregulation remains largely unknown. Here we identify two prevalent RPG promoter types, both characterized by upstream binding of the general transcription factor (TF) Rap1 followed by the RPG-specific Fhl1/Ifh1 pair, with one type also binding the HMG-B protein Hmo1. We show that the regulatory properties of the two promoter types are remarkably similar, suggesting that they are determined to a large extent by Rap1 and the Fhl1/Ifh1 pair. Rapid depletion experiments allowed us to define a hierarchy of TF binding in which Rap1 acts as a pioneer factor required for binding of all other TFs. We also uncovered unexpected features underlying recruitment of Fhl1, whose forkhead DNA-binding domain is not required for binding at most promoters, and Hmo1, whose binding is supported by repeated motifs. Finally, we describe unusually micrococcal nuclease (MNase)-sensitive nucleosomes at all RPG promoters, located between the canonical +1 and -1 nucleosomes, which coincide with sites of Fhl1/Ifh1 and Hmo1 binding. We speculate that these "fragile" nucleosomes play an important role in regulating RPG transcriptional output.
Keywords: Rap1; fragile nucleosome; ribosomal protein gene; transcription; yeast.
© 2014 Knight et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Similar articles
-
An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae.Mol Cell Biol. 2006 May;26(9):3672-9. doi: 10.1128/MCB.26.9.3672-3679.2006. Mol Cell Biol. 2006. PMID: 16612005 Free PMC article.
-
Fine-structure analysis of ribosomal protein gene transcription.Mol Cell Biol. 2006 Jul;26(13):4853-62. doi: 10.1128/MCB.02367-05. Mol Cell Biol. 2006. PMID: 16782874 Free PMC article.
-
Promoter architecture and transcriptional regulation of Abf1-dependent ribosomal protein genes in Saccharomyces cerevisiae.Nucleic Acids Res. 2016 Jul 27;44(13):6113-26. doi: 10.1093/nar/gkw194. Epub 2016 Mar 25. Nucleic Acids Res. 2016. PMID: 27016735 Free PMC article.
-
Transcriptional control of ribosome biogenesis in yeast: links to growth and stress signals.Biochem Soc Trans. 2021 Aug 27;49(4):1589-1599. doi: 10.1042/BST20201136. Biochem Soc Trans. 2021. PMID: 34240738 Free PMC article. Review.
-
Physiological function of FKBP12, a primary target of rapamycin/FK506: a newly identified role in transcription of ribosomal protein genes in yeast.Curr Genet. 2021 Jun;67(3):383-388. doi: 10.1007/s00294-020-01142-3. Epub 2021 Jan 12. Curr Genet. 2021. PMID: 33438053 Review.
Cited by
-
Translational Capacity of a Cell Is Determined during Transcription Elongation via the Ccr4-Not Complex.Cell Rep. 2016 May 24;15(8):1782-94. doi: 10.1016/j.celrep.2016.04.055. Epub 2016 May 12. Cell Rep. 2016. PMID: 27184853 Free PMC article.
-
Alignment and quantification of ChIP-exo crosslinking patterns reveal the spatial organization of protein-DNA complexes.Nucleic Acids Res. 2020 Nov 18;48(20):11215-11226. doi: 10.1093/nar/gkaa618. Nucleic Acids Res. 2020. PMID: 32747934 Free PMC article.
-
Contribution of Sequence Motif, Chromatin State, and DNA Structure Features to Predictive Models of Transcription Factor Binding in Yeast.PLoS Comput Biol. 2015 Aug 20;11(8):e1004418. doi: 10.1371/journal.pcbi.1004418. eCollection 2015 Aug. PLoS Comput Biol. 2015. PMID: 26291518 Free PMC article.
-
MNase titration reveals differences between nucleosome occupancy and chromatin accessibility.Nat Commun. 2016 May 6;7:11485. doi: 10.1038/ncomms11485. Nat Commun. 2016. PMID: 27151365 Free PMC article.
-
Fpr1, a primary target of rapamycin, functions as a transcription factor for ribosomal protein genes cooperatively with Hmo1 in Saccharomyces cerevisiae.PLoS Genet. 2020 Jun 30;16(6):e1008865. doi: 10.1371/journal.pgen.1008865. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32603360 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
