Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism
- PMID: 25083875
- PMCID: PMC5472354
- DOI: 10.1016/j.cell.2014.06.050
Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism
Abstract
Circadian rhythms are intimately linked to cellular metabolism. Specifically, the NAD(+)-dependent deacetylase SIRT1, the founding member of the sirtuin family, contributes to clock function. Whereas SIRT1 exhibits diversity in deacetylation targets and subcellular localization, SIRT6 is the only constitutively chromatin-associated sirtuin and is prominently present at transcriptionally active genomic loci. Comparison of the hepatic circadian transcriptomes reveals that SIRT6 and SIRT1 separately control transcriptional specificity and therefore define distinctly partitioned classes of circadian genes. SIRT6 interacts with CLOCK:BMAL1 and, differently from SIRT1, governs their chromatin recruitment to circadian gene promoters. Moreover, SIRT6 controls circadian chromatin recruitment of SREBP-1, resulting in the cyclic regulation of genes implicated in fatty acid and cholesterol metabolism. This mechanism parallels a phenotypic disruption in fatty acid metabolism in SIRT6 null mice as revealed by circadian metabolome analyses. Thus, genomic partitioning by two independent sirtuins contributes to differential control of circadian metabolism.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

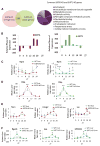
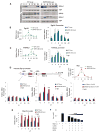
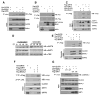
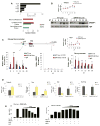
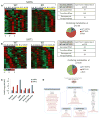
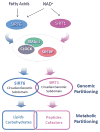
Comment in
-
Physiology. Partitioning the circadian clock.Science. 2014 Sep 5;345(6201):1122-3. doi: 10.1126/science.1259601. Science. 2014. PMID: 25190781 Free PMC article. No abstract available.
Similar articles
-
The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control.Cell. 2008 Jul 25;134(2):329-40. doi: 10.1016/j.cell.2008.07.002. Cell. 2008. PMID: 18662547 Free PMC article.
-
Sirt6 deacetylase activity regulates circadian rhythms via Per2.Biochem Biophys Res Commun. 2019 Apr 2;511(2):234-238. doi: 10.1016/j.bbrc.2019.01.143. Epub 2019 Feb 16. Biochem Biophys Res Commun. 2019. PMID: 30782483
-
Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1.Proc Natl Acad Sci U S A. 2013 Feb 26;110(9):3333-8. doi: 10.1073/pnas.1214266110. Epub 2013 Jan 22. Proc Natl Acad Sci U S A. 2013. PMID: 23341587 Free PMC article.
-
Coupling circadian rhythms of metabolism and chromatin remodelling.Diabetes Obes Metab. 2015 Sep;17 Suppl 1(0 1):17-22. doi: 10.1111/dom.12509. Diabetes Obes Metab. 2015. PMID: 26332964 Free PMC article. Review.
-
The Clock-NAD+ -Sirtuin connection in nonalcoholic fatty liver disease.J Cell Physiol. 2022 Aug;237(8):3164-3180. doi: 10.1002/jcp.30772. Epub 2022 May 26. J Cell Physiol. 2022. PMID: 35616339 Review.
Cited by
-
Simulated Night-Shift Schedule Disrupts the Plasma Lipidome and Reveals Early Markers of Cardiovascular Disease Risk.Nat Sci Sleep. 2022 May 21;14:981-994. doi: 10.2147/NSS.S363437. eCollection 2022. Nat Sci Sleep. 2022. PMID: 35645584 Free PMC article.
-
Inhibition of AKT induces p53/SIRT6/PARP1-dependent parthanatos to suppress tumor growth.Cell Commun Signal. 2022 Jun 17;20(1):93. doi: 10.1186/s12964-022-00897-1. Cell Commun Signal. 2022. PMID: 35715817 Free PMC article.
-
Regulation of circadian clock transcriptional output by CLOCK:BMAL1.PLoS Genet. 2018 Jan 4;14(1):e1007156. doi: 10.1371/journal.pgen.1007156. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29300726 Free PMC article.
-
Sirt6 Suppresses High Glucose-Induced Mitochondrial Dysfunction and Apoptosis in Podocytes through AMPK Activation.Int J Biol Sci. 2019 Jan 24;15(3):701-713. doi: 10.7150/ijbs.29323. eCollection 2019. Int J Biol Sci. 2019. PMID: 30745856 Free PMC article.
-
Experimental design and power calculation in omics circadian rhythmicity detection using the cosinor model.Stat Med. 2023 Aug 15;42(18):3236-3258. doi: 10.1002/sim.9803. Epub 2023 Jun 2. Stat Med. 2023. PMID: 37265194 Free PMC article.
References
-
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. - PubMed
-
- Bennett MK, Lopez JM, Sanchez HB, Osborne TF. Sterol regulation of fatty acid synthase promoter. Coordinate feedback regulation of two major lipid pathways. J Biol Chem. 1995;270:25578–25583. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

