Eradication of metastatic melanoma through cooperative expression of RNA-based HDAC1 inhibitor and p73 by oncolytic adenovirus
- PMID: 25071017
- PMCID: PMC4171600
- DOI: 10.18632/oncotarget.1839
Eradication of metastatic melanoma through cooperative expression of RNA-based HDAC1 inhibitor and p73 by oncolytic adenovirus
Abstract
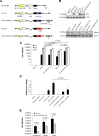
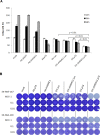
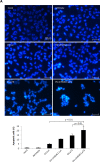
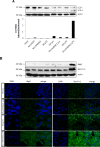
Malignant melanoma is a highly aggressive cancer that retains functional p53 and p73, and drug unresponsiveness largely depends on defects in death pathways after epigenetic gene silencing in conjunction with an imbalanced p73/DNp73 ratio. We constructed oncolytic viruses armed with an inhibitor of deacetylation and/or p73 to specifically target metastatic cancer. Arming of the viruses is aimed at lifting epigenetic blockage and re-opening apoptotic programs in a staggered manner enabling both, efficient virus replication and balanced destruction of target cells through apoptosis. Our results showed that cooperative expression of shHDAC1 and p73 efficiently enhances apoptosis induction and autophagy of infected cells which reinforces progeny production. In vitro analyses revealed 100% cytotoxicity after infecting cells with OV.shHDAC1.p73 at a lower virus dose compared to control viruses. Intriguingly, OV.shHDAC1.p73 acts as a potent inhibitor of highly metastatic xenograft tumors in vivo. Tumor expansion was significantly reduced after intratumoral injection of 3 x 10⁸ PFU of either OV.shHDAC1 or OV.p73 and, most important, complete regression could be achieved in 100 % of tumors treated with OV.shHDAC1.p73. Our results point out that the combination of high replication capacity and simultaneous restoration of cell death routes significantly enhance antitumor activity.
Figures

Similar articles
-
Silencing of Mcl-1 overcomes resistance of melanoma cells against TRAIL-armed oncolytic adenovirus by enhancement of apoptosis.J Mol Med (Berl). 2021 Sep;99(9):1279-1291. doi: 10.1007/s00109-021-02081-3. Epub 2021 May 24. J Mol Med (Berl). 2021. PMID: 34028599 Free PMC article.
-
E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry.Cell Cycle. 2012 Aug 15;11(16):3067-78. doi: 10.4161/cc.21476. Epub 2012 Aug 8. Cell Cycle. 2012. PMID: 22871739 Free PMC article.
-
Adenovirus-mediated TA-p73beta gene transfer increases chemosensitivity of human malignant melanomas.Apoptosis. 2006 Feb;11(2):235-43. doi: 10.1007/s10495-006-3407-0. Apoptosis. 2006. PMID: 16502261
-
Oncolytic adenoviruses for the treatment of brain tumors.Curr Opin Mol Ther. 2010 Oct;12(5):530-7. Curr Opin Mol Ther. 2010. PMID: 20886384 Review.
-
Recent advances in oncolytic virus-based cancer therapy.Virus Res. 2019 Sep;270:197675. doi: 10.1016/j.virusres.2019.197675. Epub 2019 Jul 25. Virus Res. 2019. PMID: 31351879 Review.
Cited by
-
Potential of histone deacetylase inhibitors in the control and regulation of prostate, breast and ovarian cancer.Front Chem. 2022 Aug 12;10:948217. doi: 10.3389/fchem.2022.948217. eCollection 2022. Front Chem. 2022. PMID: 36034650 Free PMC article. Review.
-
Role of Histone Deacetylases in Carcinogenesis: Potential Role in Cholangiocarcinoma.Cells. 2020 Mar 23;9(3):780. doi: 10.3390/cells9030780. Cells. 2020. PMID: 32210140 Free PMC article. Review.
-
BET and BRAF inhibitors act synergistically against BRAF-mutant melanoma.Cancer Med. 2016 Jun;5(6):1183-93. doi: 10.1002/cam4.667. Epub 2016 May 11. Cancer Med. 2016. PMID: 27169980 Free PMC article.
-
Delivery of cancer therapies by synthetic and bio-inspired nanovectors.Mol Cancer. 2021 Mar 24;20(1):55. doi: 10.1186/s12943-021-01346-2. Mol Cancer. 2021. PMID: 33761944 Free PMC article. Review.
-
The Role of Regional Therapies for in-Transit Melanoma in the Era of Improved Systemic Options.Cancers (Basel). 2015 Jul 1;7(3):1154-77. doi: 10.3390/cancers7030830. Cancers (Basel). 2015. PMID: 26140669 Free PMC article. Review.
References
-
- Garbe C, Peris K, Hauschild A, Saiag P, Middleton M, Spatz A, Grob JJ, Malvehy J, Newton-Bishop J, Stratigos A, Pehamberger H, Eggermont AM. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline – Update 2012. Eur J Cancer. 2012;48:2375–90. - PubMed
-
- Pützer BM, Steder M, Alla V. Predicting and preventing melanoma invasiveness: advances in clarifying E2F1 function. Expert Rev Anticancer Ther. 2010;10:1707–20. - PubMed
-
- Dahl C, Guldberg P. The genome and epigenome of malignant melanoma. APMIS. 2007;115:1161–76. - PubMed
-
- Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

