Type II transmembrane serine protease gene variants associate with breast cancer
- PMID: 25029565
- PMCID: PMC4100901
- DOI: 10.1371/journal.pone.0102519
Type II transmembrane serine protease gene variants associate with breast cancer
Abstract
Type II transmembrane serine proteases (TTSPs) are related to tumor growth, invasion, and metastasis in cancer. Genetic variants in these genes may alter their function, leading to cancer onset and progression, and affect patient outcome. Here, 464 breast cancer cases and 370 controls were genotyped for 82 single-nucleotide polymorphisms covering eight genes. Association of the genotypes was estimated against breast cancer risk, breast cancer-specific survival, and survival in different treatment groups, and clinicopathological variables. SNPs in TMPRSS3 (rs3814903 and rs11203200), TMPRSS7 (rs1844925), and HGF (rs5745752) associated significantly with breast cancer risk (Ptrend = 0.008-0.042). SNPs in TMPRSS1 (rs12151195 and rs12461158), TMPRSS2 (rs2276205), TMPRSS3 (rs3814903), and TMPRSS7 (rs2399403) associated with prognosis (P = 0.004-0.046). When estimating the combined effect of the variants, the risk of breast cancer was higher with 4-5 alleles present compared to 0-2 alleles (P = 0.0001; OR, 2.34; 95% CI, 1.39-3.94). Women with 6-8 survival-associating alleles had a 3.3 times higher risk of dying of breast cancer compared to women with 1-3 alleles (P = 0.001; HR, 3.30; 95% CI, 1.58-6.88). The results demonstrate the combined effect of variants in TTSPs and their related genes in breast cancer risk and patient outcome. Functional analysis of these variants will lead to further understanding of this gene family, which may improve individualized risk estimation and development of new strategies for treatment of breast cancer.
Conflict of interest statement
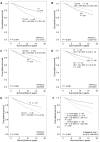
Figures
Similar articles
-
Low expression levels of hepsin and TMPRSS3 are associated with poor breast cancer survival.BMC Cancer. 2015 May 27;15:431. doi: 10.1186/s12885-015-1440-5. BMC Cancer. 2015. PMID: 26014348 Free PMC article.
-
Matriptase-2 gene (TMPRSS6) variants associate with breast cancer survival, and reduced expression is related to triple-negative breast cancer.Int J Cancer. 2013 Nov 15;133(10):2334-40. doi: 10.1002/ijc.28254. Epub 2013 Jun 4. Int J Cancer. 2013. PMID: 23649491
-
Genetic Polymorphism of SUMO-Specific Cysteine Proteases - SENP1 and SENP2 in Breast Cancer.Pathol Oncol Res. 2016 Oct;22(4):817-23. doi: 10.1007/s12253-016-0064-7. Epub 2016 May 13. Pathol Oncol Res. 2016. PMID: 27178176 Free PMC article.
-
Preliminary investigation of MMP8 (rs11225395) and MMP9 (rs3787268) polymorphisms association with breast cancer risk in pashtun women of Pakistan.Mol Biol Rep. 2024 Oct 3;51(1):1034. doi: 10.1007/s11033-024-09968-7. Mol Biol Rep. 2024. PMID: 39361067
-
Analysis of functional germline variants in APOBEC3 and driver genes on breast cancer risk in Moroccan study population.BMC Cancer. 2016 Feb 26;16:165. doi: 10.1186/s12885-016-2210-8. BMC Cancer. 2016. PMID: 26920143 Free PMC article.
Cited by
-
COVID-19 Mechanisms in the Human Body-What We Know So Far.Front Immunol. 2021 Nov 1;12:693938. doi: 10.3389/fimmu.2021.693938. eCollection 2021. Front Immunol. 2021. PMID: 34790191 Free PMC article. Review.
-
Oncogenic Notch Promotes Long-Range Regulatory Interactions within Hyperconnected 3D Cliques.Mol Cell. 2019 Mar 21;73(6):1174-1190.e12. doi: 10.1016/j.molcel.2019.01.006. Epub 2019 Feb 7. Mol Cell. 2019. PMID: 30745086 Free PMC article.
-
Semantic and Population Analysis of the Genetic Targets Related to COVID-19 and Its Association with Genes and Diseases.Adv Exp Med Biol. 2023;1423:59-78. doi: 10.1007/978-3-031-31978-5_6. Adv Exp Med Biol. 2023. PMID: 37525033
-
Dexmedetomidine promotes breast cancer cell migration through Rab11-mediated secretion of exosomal TMPRSS2.Ann Transl Med. 2020 Apr;8(8):531. doi: 10.21037/atm.2020.04.28. Ann Transl Med. 2020. PMID: 32411754 Free PMC article.
-
Low expression levels of hepsin and TMPRSS3 are associated with poor breast cancer survival.BMC Cancer. 2015 May 27;15:431. doi: 10.1186/s12885-015-1440-5. BMC Cancer. 2015. PMID: 26014348 Free PMC article.
References
-
- Stratton MR, Rahman N (2008) The emerging landscape of breast cancer susceptibility. Nat Genet 40: 17–22. - PubMed
-
- Burton H, Chowdhury S, Dent T, Hall A, Pashayan N, et al. (2013) Public health implications from COGS and potential for risk stratification and screening. Nat Genet 45: 349–351. - PubMed
-
- Hooper JD, Clements JA, Quigley JP, Antalis TM (2001) Type II transmembrane serine proteases. insights into an emerging class of cell surface proteolytic enzymes. J Biol Chem 276: 857–860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

