Transcriptional landscape of repetitive elements in normal and cancer human cells
- PMID: 25012247
- PMCID: PMC4122776
- DOI: 10.1186/1471-2164-15-583
Transcriptional landscape of repetitive elements in normal and cancer human cells
Abstract
Background: Repetitive elements comprise at least 55% of the human genome with more recent estimates as high as two-thirds. Most of these elements are retrotransposons, DNA sequences that can insert copies of themselves into new genomic locations by a "copy and paste" mechanism. These mobile genetic elements play important roles in shaping genomes during evolution, and have been implicated in the etiology of many human diseases. Despite their abundance and diversity, few studies investigated the regulation of endogenous retrotransposons at the genome-wide scale, primarily because of the technical difficulties of uniquely mapping high-throughput sequencing reads to repetitive DNA.
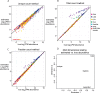
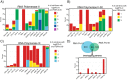
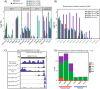
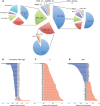
Results: Here we develop a new computational method called RepEnrich to study genome-wide transcriptional regulation of repetitive elements. We show that many of the Long Terminal Repeat retrotransposons in humans are transcriptionally active in a cell line-specific manner. Cancer cell lines display increased RNA Polymerase II binding to retrotransposons than cell lines derived from normal tissue. Consistent with increased transcriptional activity of retrotransposons in cancer cells we found significantly higher levels of L1 retrotransposon RNA expression in prostate tumors compared to normal-matched controls.
Conclusions: Our results support increased transcription of retrotransposons in transformed cells, which may explain the somatic retrotransposition events recently reported in several types of cancers.
Figures


Similar articles
-
The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-insertion Sequence Biases and Post-insertion Selection.Mol Cell. 2019 May 2;74(3):555-570.e7. doi: 10.1016/j.molcel.2019.02.036. Epub 2019 Apr 4. Mol Cell. 2019. PMID: 30956044
-
Analysis of RNA-Seq Data Using TEtranscripts.Methods Mol Biol. 2018;1751:153-167. doi: 10.1007/978-1-4939-7710-9_11. Methods Mol Biol. 2018. PMID: 29508296
-
An update on post-transcriptional regulation of retrotransposons.FEBS Lett. 2023 Feb;597(3):380-406. doi: 10.1002/1873-3468.14551. Epub 2022 Dec 12. FEBS Lett. 2023. PMID: 36460901 Review.
-
Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes.Science. 2014 Aug 1;345(6196):1251343. doi: 10.1126/science.1251343. Science. 2014. PMID: 25082706 Free PMC article.
-
Crossing the LINE Toward Genomic Instability: LINE-1 Retrotransposition in Cancer.Front Chem. 2015 Dec 16;3:68. doi: 10.3389/fchem.2015.00068. eCollection 2015. Front Chem. 2015. PMID: 26734601 Free PMC article. Review.
Cited by
-
Pericentromeric Satellite III transcripts induce etoposide resistance.Cell Death Dis. 2021 May 24;12(6):530. doi: 10.1038/s41419-021-03810-9. Cell Death Dis. 2021. PMID: 34031359 Free PMC article.
-
Co-option of the lineage-specific LAVA retrotransposon in the gibbon genome.Proc Natl Acad Sci U S A. 2020 Aug 11;117(32):19328-19338. doi: 10.1073/pnas.2006038117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690705 Free PMC article.
-
Expression of evolutionarily novel genes in tumors.Infect Agent Cancer. 2016 Jul 19;11:34. doi: 10.1186/s13027-016-0077-6. eCollection 2016. Infect Agent Cancer. 2016. PMID: 27437030 Free PMC article. Review.
-
HMGA2 directly mediates chromatin condensation in association with neuronal fate regulation.Nat Commun. 2023 Oct 12;14(1):6420. doi: 10.1038/s41467-023-42094-9. Nat Commun. 2023. PMID: 37828010 Free PMC article.
-
Landscape of Transposable Elements Focusing on the B Chromosome of the Cichlid Fish Astatotilapia latifasciata.Genes (Basel). 2018 May 23;9(6):269. doi: 10.3390/genes9060269. Genes (Basel). 2018. PMID: 29882892 Free PMC article.
References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

