Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon
- PMID: 25005358
- PMCID: PMC4268115
- DOI: 10.1038/mi.2014.58
Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon
Abstract
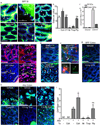
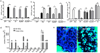
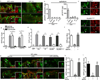
The delivery of luminal substances across the intestinal epithelium to the immune system is a critical event in immune surveillance, resulting in tolerance to dietary antigens and immunity to pathogens. How this process is regulated is largely unknown. Recently goblet cell-associated antigen passages (GAPs) were identified as a pathway delivering luminal antigens to underlying lamina propria (LP) dendritic cells in the steady state. Here, we demonstrate that goblet cells (GCs) form GAPs in response to acetylcholine (ACh) acting on muscarinic ACh receptor 4. GAP formation in the small intestine was regulated at the level of ACh production, as GCs rapidly formed GAPs in response to ACh analogs. In contrast, colonic GAP formation was regulated at the level of GC responsiveness to ACh. Myd88-dependent microbial sensing by colonic GCs inhibited the ability of colonic GCs to respond to Ach to form GAPs and deliver luminal antigens to colonic LP-antigen-presenting cells (APCs). Disruption of GC microbial sensing in the setting of an intact gut microbiota opened colonic GAPs, and resulted in recruitment of neutrophils and APCs and production of inflammatory cytokines. Thus GC intrinsic sensing of the microbiota has a critical role regulating the exposure of the colonic immune system to luminal substances.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Goblet cells: multifaceted players in immunity at mucosal surfaces.Mucosal Immunol. 2018 Nov;11(6):1551-1557. doi: 10.1038/s41385-018-0039-y. Epub 2018 Jun 4. Mucosal Immunol. 2018. PMID: 29867079 Free PMC article. Review.
-
In vivo labeling of epithelial cell-associated antigen passages in the murine intestine.Lab Anim (NY). 2020 Mar;49(3):79-88. doi: 10.1038/s41684-019-0438-z. Epub 2020 Feb 10. Lab Anim (NY). 2020. PMID: 32042160 Free PMC article.
-
Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine.Nature. 2012 Mar 14;483(7389):345-9. doi: 10.1038/nature10863. Nature. 2012. PMID: 22422267 Free PMC article.
-
Goblet cell associated antigen passages support the induction and maintenance of oral tolerance.Mucosal Immunol. 2020 Mar;13(2):271-282. doi: 10.1038/s41385-019-0240-7. Epub 2019 Dec 9. Mucosal Immunol. 2020. PMID: 31819172 Free PMC article.
-
Transepithelial antigen delivery in the small intestine: different paths, different outcomes.Curr Opin Gastroenterol. 2013 Mar;29(2):112-8. doi: 10.1097/MOG.0b013e32835cf1cd. Curr Opin Gastroenterol. 2013. PMID: 23380572 Free PMC article. Review.
Cited by
-
Contribution of mucus concentration and secreted mucins Muc5ac and Muc5b to the pathogenesis of muco-obstructive lung disease.Mucosal Immunol. 2017 Mar;10(2):395-407. doi: 10.1038/mi.2016.63. Epub 2016 Jul 20. Mucosal Immunol. 2017. PMID: 27435107 Free PMC article.
-
The renaissance of oral tolerance: merging tradition and new insights.Nat Rev Immunol. 2025 Jan;25(1):42-56. doi: 10.1038/s41577-024-01077-7. Epub 2024 Sep 6. Nat Rev Immunol. 2025. PMID: 39242920 Review.
-
Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon.Proc Natl Acad Sci U S A. 2016 Sep 13;113(37):E5399-407. doi: 10.1073/pnas.1607327113. Epub 2016 Aug 29. Proc Natl Acad Sci U S A. 2016. PMID: 27573849 Free PMC article.
-
"Iron triangle" of regulating the uterine microecology: Endometrial microbiota, immunity and endometrium.Front Immunol. 2022 Aug 9;13:928475. doi: 10.3389/fimmu.2022.928475. eCollection 2022. Front Immunol. 2022. PMID: 36016947 Free PMC article. Review.
-
Goblet cells: multifaceted players in immunity at mucosal surfaces.Mucosal Immunol. 2018 Nov;11(6):1551-1557. doi: 10.1038/s41385-018-0039-y. Epub 2018 Jun 4. Mucosal Immunol. 2018. PMID: 29867079 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK064798/DK/NIDDK NIH HHS/United States
- P30AR048335/AR/NIAMS NIH HHS/United States
- DK097317-RDN/DK/NIDDK NIH HHS/United States
- P30 CA91842/CA/NCI NIH HHS/United States
- R01 DK097317/DK/NIDDK NIH HHS/United States
- AI095550-RDN/AI/NIAID NIH HHS/United States
- R21 AI083538/AI/NIAID NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- DK097893-KAK/DK/NIDDK NIH HHS/United States
- U01 AI095550/AI/NIAID NIH HHS/United States
- F32 DK097893/DK/NIDDK NIH HHS/United States
- R01 AI077600/AI/NIAID NIH HHS/United States
- P30 AR048335/AR/NIAMS NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- DK064798-RDN/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

