TOR complex 2-Ypk1 signaling is an essential positive regulator of the general amino acid control response and autophagy
- PMID: 25002487
- PMCID: PMC4115538
- DOI: 10.1073/pnas.1406305111
TOR complex 2-Ypk1 signaling is an essential positive regulator of the general amino acid control response and autophagy
Abstract
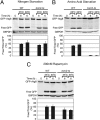
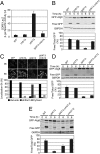
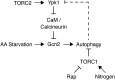
The highly conserved Target of Rapamycin (TOR) kinase is a central regulator of cell growth and metabolism in response to nutrient availability. TOR functions in two structurally and functionally distinct complexes, TOR Complex 1 (TORC1) and TOR Complex 2 (TORC2). Through TORC1, TOR negatively regulates autophagy, a conserved process that functions in quality control and cellular homeostasis and, in this capacity, is part of an adaptive nutrient deprivation response. Here we demonstrate that during amino acid starvation TOR also operates independently as a positive regulator of autophagy through the conserved TORC2 and its downstream target protein kinase, Ypk1. Under these conditions, TORC2-Ypk1 signaling negatively regulates the Ca(2+)/calmodulin-dependent phosphatase, calcineurin, to enable the activation of the amino acid-sensing eIF2α kinase, Gcn2, and to promote autophagy. Our work reveals that the TORC2 pathway regulates autophagy in an opposing manner to TORC1 to provide a tunable response to cellular metabolic status.
Keywords: Atg8; Gcn4.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Calcium channel regulator Mid1 links TORC2-mediated changes in mitochondrial respiration to autophagy.J Cell Biol. 2016 Dec 19;215(6):779-788. doi: 10.1083/jcb.201605030. Epub 2016 Nov 29. J Cell Biol. 2016. PMID: 27899413 Free PMC article.
-
A role for TOR complex 2 signaling in promoting autophagy.Autophagy. 2014;10(11):2085-6. doi: 10.4161/auto.36262. Autophagy. 2014. PMID: 25426890 Free PMC article.
-
TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids.Elife. 2014 Oct 3;3:e03779. doi: 10.7554/eLife.03779. Elife. 2014. PMID: 25279700 Free PMC article.
-
Evolutionarily conserved regulation of TOR signalling.J Biochem. 2013 Jul;154(1):1-10. doi: 10.1093/jb/mvt047. Epub 2013 May 21. J Biochem. 2013. PMID: 23698095 Review.
-
Regulation of TORC2 function and localization by Rab5 GTPases in Saccharomyces cerevisiae.Cell Cycle. 2019 May;18(10):1084-1094. doi: 10.1080/15384101.2019.1616999. Epub 2019 May 15. Cell Cycle. 2019. PMID: 31068077 Free PMC article. Review.
Cited by
-
Calcium channel regulator Mid1 links TORC2-mediated changes in mitochondrial respiration to autophagy.J Cell Biol. 2016 Dec 19;215(6):779-788. doi: 10.1083/jcb.201605030. Epub 2016 Nov 29. J Cell Biol. 2016. PMID: 27899413 Free PMC article.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition).Autophagy. 2016;12(1):1-222. doi: 10.1080/15548627.2015.1100356. Autophagy. 2016. PMID: 26799652 Free PMC article. No abstract available.
-
Autophagy modulation: a potential therapeutic approach in cardiac hypertrophy.Am J Physiol Heart Circ Physiol. 2017 Aug 1;313(2):H304-H319. doi: 10.1152/ajpheart.00145.2017. Epub 2017 Jun 2. Am J Physiol Heart Circ Physiol. 2017. PMID: 28576834 Free PMC article. Review.
-
The mTORC1/4E-BP pathway coordinates hemoglobin production with L-leucine availability.Sci Signal. 2015 Apr 14;8(372):ra34. doi: 10.1126/scisignal.aaa5903. Sci Signal. 2015. PMID: 25872869 Free PMC article.
-
A Signaling Lipid Associated with Alzheimer's Disease Promotes Mitochondrial Dysfunction.Sci Rep. 2016 Jan 13;6:19332. doi: 10.1038/srep19332. Sci Rep. 2016. PMID: 26757638 Free PMC article.
References
-
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1-2):169–174. - PubMed
-
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280(36):31582–31586. - PubMed
-
- Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12(9):842–846. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

