Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility
- PMID: 24997565
- PMCID: PMC4140783
- DOI: 10.1038/ni.2937
Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility
Abstract
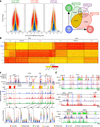
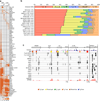
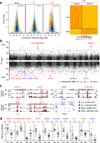
A characteristic feature of asthma is the aberrant accumulation, differentiation or function of memory CD4(+) T cells that produce type 2 cytokines (TH2 cells). By mapping genome-wide histone modification profiles for subsets of T cells isolated from peripheral blood of healthy and asthmatic individuals, we identified enhancers with known and potential roles in the normal differentiation of human TH1 cells and TH2 cells. We discovered disease-specific enhancers in T cells that differ between healthy and asthmatic individuals. Enhancers that gained the histone H3 Lys4 dimethyl (H3K4me2) mark during TH2 cell development showed the highest enrichment for asthma-associated single nucleotide polymorphisms (SNPs), which supported a pathogenic role for TH2 cells in asthma. In silico analysis of cell-specific enhancers revealed transcription factors, microRNAs and genes potentially linked to human TH2 cell differentiation. Our results establish the feasibility and utility of enhancer profiling in well-defined populations of specialized cell types involved in disease pathogenesis.
Figures



Comment in
-
Enhancing the understanding of asthma.Nat Immunol. 2014 Aug;15(8):701-3. doi: 10.1038/ni.2946. Nat Immunol. 2014. PMID: 25045871 Free PMC article. No abstract available.
Similar articles
-
The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells.Immunity. 2013 Nov 14;39(5):819-32. doi: 10.1016/j.immuni.2013.09.012. Immunity. 2013. PMID: 24238339
-
[Imbalanced T cell-specific transcription factors T-bet and GATA-3 contributes to type 2 T helper cell polarization in asthmatic patients].Zhonghua Jie He He Hu Xi Za Zhi. 2004 Jun;27(6):398-402. Zhonghua Jie He He Hu Xi Za Zhi. 2004. PMID: 15256090 Chinese.
-
Fine particulate matter induces airway inflammation by disturbing the balance between Th1/Th2 and regulation of GATA3 and Runx3 expression in BALB/c mice.Mol Med Rep. 2021 May;23(5):378. doi: 10.3892/mmr.2021.12017. Epub 2021 Mar 24. Mol Med Rep. 2021. PMID: 33760131 Free PMC article.
-
[Th1/Th2 differentiation].Nihon Rinsho. 2005 Apr;63 Suppl 4:355-63. Nihon Rinsho. 2005. PMID: 15861681 Review. Japanese. No abstract available.
-
'All things considered': transcriptional regulation of T helper type 2 cell differentiation from precursor to effector activation.Immunology. 2013 Sep;140(1):31-8. doi: 10.1111/imm.12121. Immunology. 2013. PMID: 23668241 Free PMC article. Review.
Cited by
-
The Role of Systems Biology in Deciphering Asthma Heterogeneity.Life (Basel). 2022 Oct 8;12(10):1562. doi: 10.3390/life12101562. Life (Basel). 2022. PMID: 36294997 Free PMC article. Review.
-
CD4+ T-cell subsets in inflammatory diseases: beyond the Th1/Th2 paradigm.Int Immunol. 2016 Apr;28(4):163-71. doi: 10.1093/intimm/dxw006. Epub 2016 Feb 12. Int Immunol. 2016. PMID: 26874355 Free PMC article. Review.
-
Regulation of human helper T cell subset differentiation by cytokines.Curr Opin Immunol. 2015 Jun;34:130-6. doi: 10.1016/j.coi.2015.03.007. Epub 2015 Apr 11. Curr Opin Immunol. 2015. PMID: 25879814 Free PMC article. Review.
-
DNA methylation signature of interleukin 1 receptor type II in asthma.Clin Epigenetics. 2015 Aug 5;7(1):80. doi: 10.1186/s13148-015-0114-0. eCollection 2015. Clin Epigenetics. 2015. PMID: 26246860 Free PMC article.
-
Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers.Immunity. 2022 Mar 8;55(3):557-574.e7. doi: 10.1016/j.immuni.2022.02.004. Immunity. 2022. PMID: 35263570 Free PMC article.
References
-
- Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. - PubMed
-
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616–623. - PubMed
-
- Kay AB. Allergy and allergic diseases. Second of two parts. N. Engl. J. Med. 2001;344:109–113. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

