Structure of Yin Yang 1 oligomers that cooperate with RuvBL1-RuvBL2 ATPases
- PMID: 24990942
- PMCID: PMC4132769
- DOI: 10.1074/jbc.M114.567040
Structure of Yin Yang 1 oligomers that cooperate with RuvBL1-RuvBL2 ATPases
Abstract
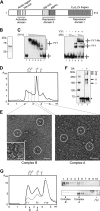
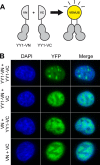
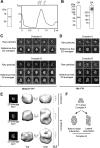
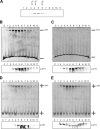
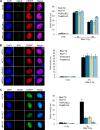
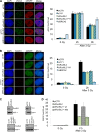
Yin Yang 1 (YY1) is a transcription factor regulating proliferation and differentiation and is involved in cancer development. Oligomers of recombinant YY1 have been observed before, but their structure and DNA binding properties are not well understood. Here we find that YY1 assembles several homo-oligomeric species built from the association of a bell-shaped dimer, a process we characterized by electron microscopy. Moreover, we find that YY1 self-association also occurs in vivo using bimolecular fluorescence complementation. Unexpectedly, these oligomers recognize several DNA substrates without the consensus sequence for YY1 in vitro, and DNA binding is enhanced in the presence of RuvBL1-RuvBL2, two essential AAA+ ATPases. YY1 oligomers bind RuvBL1-RuvBL2 hetero-oligomeric complexes, but YY1 interacts preferentially with RuvBL1. Collectively, these findings suggest that YY1-RuvBL1-RuvBL2 complexes could contribute to functions beyond transcription, and we show that YY1 and the ATPase activity of RuvBL2 are required for RAD51 foci formation during homologous recombination.
Keywords: ATPase; DNA Repair; Electron Microscopy (EM); RuvBL1; RuvBL2; Single Particle Analysis; Structural Biology; Transcription Factor; YY1.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Regulation of RUVBL1-RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM.Elife. 2020 Nov 18;9:e63042. doi: 10.7554/eLife.63042. Elife. 2020. PMID: 33205750 Free PMC article.
-
RUVBL1/RUVBL2 ATPase Activity Drives PAQosome Maturation, DNA Replication and Radioresistance in Lung Cancer.Cell Chem Biol. 2020 Jan 16;27(1):105-121.e14. doi: 10.1016/j.chembiol.2019.12.005. Epub 2019 Dec 26. Cell Chem Biol. 2020. PMID: 31883965 Free PMC article.
-
Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.Sci Adv. 2019 May 1;5(5):eaaw1616. doi: 10.1126/sciadv.aaw1616. eCollection 2019 May. Sci Adv. 2019. PMID: 31049401 Free PMC article.
-
RUVBL1-RUVBL2 AAA-ATPase: a versatile scaffold for multiple complexes and functions.Curr Opin Struct Biol. 2021 Apr;67:78-85. doi: 10.1016/j.sbi.2020.08.010. Epub 2020 Oct 28. Curr Opin Struct Biol. 2021. PMID: 33129013 Review.
-
RPAP3 C-Terminal Domain: A Conserved Domain for the Assembly of R2TP Co-Chaperone Complexes.Cells. 2020 May 6;9(5):1139. doi: 10.3390/cells9051139. Cells. 2020. PMID: 32384603 Free PMC article. Review.
Cited by
-
Mechanisms of Enhancer-Promoter Interactions in Higher Eukaryotes.Int J Mol Sci. 2021 Jan 12;22(2):671. doi: 10.3390/ijms22020671. Int J Mol Sci. 2021. PMID: 33445415 Free PMC article. Review.
-
The AAA+ proteins Pontin and Reptin enter adult age: from understanding their basic biology to the identification of selective inhibitors.Front Mol Biosci. 2015 May 5;2:17. doi: 10.3389/fmolb.2015.00017. eCollection 2015. Front Mol Biosci. 2015. PMID: 25988184 Free PMC article.
-
Beyond Transcription: Roles of Transcription Factors in Pre-mRNA Splicing.Chem Rev. 2018 Apr 25;118(8):4339-4364. doi: 10.1021/acs.chemrev.7b00470. Epub 2017 Dec 18. Chem Rev. 2018. PMID: 29251915 Free PMC article. Review.
-
The Role of Protein Arginine Methyltransferases in DNA Damage Response.Int J Mol Sci. 2022 Aug 29;23(17):9780. doi: 10.3390/ijms23179780. Int J Mol Sci. 2022. PMID: 36077176 Free PMC article. Review.
-
CryoEM of RUVBL1-RUVBL2-ZNHIT2, a complex that interacts with pre-mRNA-processing-splicing factor 8.Nucleic Acids Res. 2022 Jan 25;50(2):1128-1146. doi: 10.1093/nar/gkab1267. Nucleic Acids Res. 2022. PMID: 34951455 Free PMC article.
References
-
- Gordon S., Akopyan G., Garban H., Bonavida B. (2006) Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25, 1125–1142 - PubMed
-
- Calame K., Atchison M. (2007) YY1 helps to bring loose ends together. Genes Dev. 21, 1145–1152 - PubMed
-
- Nicholson S., Whitehouse H., Naidoo K., Byers R. J. (2011) Yin Yang 1 in human cancer. Crit. Rev. Oncog. 16, 245–260 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

