Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice
- PMID: 24979782
- PMCID: PMC4084454
- DOI: 10.1073/pnas.1402448111
Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice
Abstract
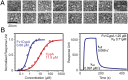
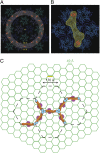
Restriction factors (RFs) form important components of host defenses to retroviral infection. The Fv1, Trim5α, and TrimCyp RFs contain N-terminal dimerization and C-terminal specificity domains that target assembled retroviral capsid (CA) proteins enclosing the viral core. However, the molecular detail of the interaction between RFs and their CA targets is unknown. Therefore, we have determined the crystal structure of the B-box and coiled-coil (BCC) region from Trim5α and used small-angle X-ray scattering to examine the solution structure of Trim5α BCC, the dimerization domain of Fv1 (Fv1Ntd), and the hybrid restriction factor Fv1Cyp comprising Fv1NtD fused to the HIV-1 binding protein Cyclophilin A (CypA). These data reveal that coiled-coil regions of Fv1 and Trim5α form extended antiparallel dimers. In Fv1Cyp, two CypA moieties are located at opposing ends, creating a molecule with a dumbbell appearance. In Trim5α, the B-boxes are located at either end of the coiled-coil, held in place by interactions with a helical motif from the L2 region of the opposing monomer. A comparative analysis of Fv1Cyp and CypA binding to a preformed HIV-1 CA lattice reveals how RF dimerization enhances the affinity of interaction through avidity effects. We conclude that the antiparallel organization of the NtD regions of Fv1 and Trim5α dimers correctly positions C-terminal specificity and N-terminal effector domains and facilitates stable binding to adjacent CA hexamers in viral cores.
Keywords: MLV; SAXS; X-ray crystallography; retrovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

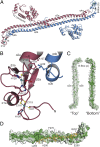
Similar articles
-
General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article.
-
The Three-Fold Axis of the HIV-1 Capsid Lattice Is the Species-Specific Binding Interface for TRIM5α.J Virol. 2018 Feb 12;92(5):e01541-17. doi: 10.1128/JVI.01541-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237846 Free PMC article.
-
A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction.J Virol. 2009 Oct;83(20):10737-51. doi: 10.1128/JVI.01307-09. Epub 2009 Aug 5. J Virol. 2009. PMID: 19656869 Free PMC article.
-
TRIM5alpha.Curr Top Microbiol Immunol. 2009;339:47-66. doi: 10.1007/978-3-642-02175-6_3. Curr Top Microbiol Immunol. 2009. PMID: 20012523 Review.
-
Retroviral restriction factors TRIM5α: therapeutic strategy to inhibit HIV-1 replication.Curr Med Chem. 2011;18(17):2649-54. doi: 10.2174/092986711795933687. Curr Med Chem. 2011. PMID: 21568899 Review.
Cited by
-
Human TRIM5α senses and restricts LINE-1 elements.Proc Natl Acad Sci U S A. 2020 Jul 28;117(30):17965-17976. doi: 10.1073/pnas.1922366117. Epub 2020 Jul 10. Proc Natl Acad Sci U S A. 2020. PMID: 32651277 Free PMC article.
-
Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway.Cell Rep. 2016 Aug 2;16(5):1315-1325. doi: 10.1016/j.celrep.2016.06.070. Epub 2016 Jul 14. Cell Rep. 2016. PMID: 27425606 Free PMC article.
-
Duplication and divergence of the retrovirus restriction gene Fv1 in Mus caroli allows protection from multiple retroviruses.PLoS Genet. 2020 Jun 11;16(6):e1008471. doi: 10.1371/journal.pgen.1008471. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32525879 Free PMC article.
-
Solution NMR structure of the TRIM21 B-box2 and identification of residues involved in its interaction with the RING domain.PLoS One. 2017 Jul 28;12(7):e0181551. doi: 10.1371/journal.pone.0181551. eCollection 2017. PLoS One. 2017. PMID: 28753623 Free PMC article.
-
TRIM5α SPRY/coiled-coil interactions optimize avid retroviral capsid recognition.PLoS Pathog. 2017 Oct 17;13(10):e1006686. doi: 10.1371/journal.ppat.1006686. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 29040325 Free PMC article.
References
-
- Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10(6):395–406. - PubMed
-
- Sanz-Ramos M, Stoye JP. Capsid-binding retrovirus restriction factors: Discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol. 2013;94(Pt 12):2587–98. - PubMed
-
- Lilly F. Fv-2: Identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J Natl Cancer Inst. 1970;45(1):163–169. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

