Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies
- PMID: 24970214
- PMCID: PMC4030991
- DOI: 10.3390/biom4010235
Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies
Abstract
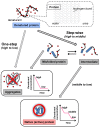
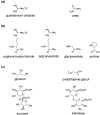
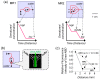
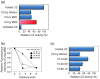
Biologically active proteins are useful for studying the biological functions of genes and for the development of therapeutic drugs and biomaterials in a biotechnology industry. Overexpression of recombinant proteins in bacteria, such as Escherichia coli, often results in the formation of inclusion bodies, which are protein aggregates with non-native conformations. As inclusion bodies contain relatively pure and intact proteins, protein refolding is an important process to obtain active recombinant proteins from inclusion bodies. However, conventional refolding methods, such as dialysis and dilution, are time consuming and, often, recovered yields of active proteins are low, and a trial-and-error process is required to achieve success. Recently, several approaches have been reported to refold these aggregated proteins into an active form. The strategies largely aim at reducing protein aggregation during the refolding procedure. This review focuses on protein refolding techniques using chemical additives and laminar flow in microfluidic chips for the efficient recovery of active proteins from inclusion bodies.
Figures

Similar articles
-
Microfluidic chips with multi-junctions: an advanced tool in recovering proteins from inclusion bodies.Bioengineered. 2015;6(1):1-4. doi: 10.4161/21655979.2014.987022. Epub 2015 Jan 7. Bioengineered. 2015. PMID: 25531187 Free PMC article.
-
[In vitro renaturation of proteins from inclusion bodies].Postepy Hig Med Dosw (Online). 2012 Jun 11;66:322-9. doi: 10.5604/17322693.999918. Postepy Hig Med Dosw (Online). 2012. PMID: 22706118 Review. Polish.
-
Refolding solubilized inclusion body proteins.Methods Enzymol. 2009;463:259-82. doi: 10.1016/S0076-6879(09)63017-2. Methods Enzymol. 2009. PMID: 19892177 Review.
-
An efficient in vitro refolding of recombinant bacterial laccase in Escherichia coli.Enzyme Microb Technol. 2013 May 10;52(6-7):325-30. doi: 10.1016/j.enzmictec.2013.03.006. Epub 2013 Mar 15. Enzyme Microb Technol. 2013. PMID: 23608500
-
Integrated refolding techniques for Schistosoma japonicum MTH1 overexpressed as inclusion bodies in Escherichia coli.Protein Expr Purif. 2012 Aug;84(2):181-7. doi: 10.1016/j.pep.2012.05.005. Epub 2012 May 26. Protein Expr Purif. 2012. PMID: 22641057
Cited by
-
Production of soluble truncated spike protein of porcine epidemic diarrhea virus from inclusion bodies of Escherichia coli through refolding.Protein Expr Purif. 2016 Oct;126:77-83. doi: 10.1016/j.pep.2016.05.018. Epub 2016 May 31. Protein Expr Purif. 2016. PMID: 27260969 Free PMC article.
-
Production of Recombinant Streptavidin and Optimization of Refolding Conditions for Recovery of Biological Activity.Rep Biochem Mol Biol. 2018 Apr;6(2):178-185. Rep Biochem Mol Biol. 2018. PMID: 29766001 Free PMC article.
-
REFOLDdb: a new and sustainable gateway to experimental protocols for protein refolding.BMC Struct Biol. 2017 Apr 24;17(1):4. doi: 10.1186/s12900-017-0074-z. BMC Struct Biol. 2017. PMID: 28438161 Free PMC article.
-
Development and Characterization of a Camelid Single Domain Antibody-Urease Conjugate That Targets Vascular Endothelial Growth Factor Receptor 2.Front Immunol. 2017 Aug 21;8:956. doi: 10.3389/fimmu.2017.00956. eCollection 2017. Front Immunol. 2017. PMID: 28871252 Free PMC article.
-
Teaching an old pET new tricks: tuning of inclusion body formation and properties by a mixed feed system in E. coli.Appl Microbiol Biotechnol. 2018 Jan;102(2):667-676. doi: 10.1007/s00253-017-8641-6. Epub 2017 Nov 20. Appl Microbiol Biotechnol. 2018. PMID: 29159587 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

