Deletion of the highly conserved N-glycan at Asn260 of HIV-1 gp120 affects folding and lysosomal degradation of gp120, and results in loss of viral infectivity
- PMID: 24967714
- PMCID: PMC4072736
- DOI: 10.1371/journal.pone.0101181
Deletion of the highly conserved N-glycan at Asn260 of HIV-1 gp120 affects folding and lysosomal degradation of gp120, and results in loss of viral infectivity
Erratum in
- PLoS One. 2014;9(10):e110202
Abstract
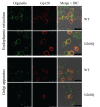
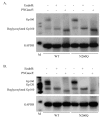
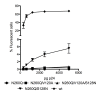
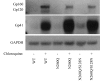
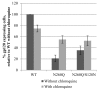
N-linked glycans covering the surface of the HIV-1 glycoprotein gp120 are of major importance for the correct folding of this glycoprotein. Of the, on average, 24 N-linked glycans present on gp120, the glycan at Asn260 was reported to be essential for the correct expression of gp120 and gp41 in the virus particle and deletion of the N260 glycan in gp120 heavily compromised virus infectivity. We show here that gp160 containing the N260Q mutation reaches the Golgi apparatus during biosynthesis. Using pulse-chase experiments with [35S] methionine/cysteine, we show that oxidative folding was slightly delayed in case of mutant N260Q gp160 and that CD4 binding was markedly compromised compared to wild-type gp160. In the search of compensatory mutations, we found a mutation in the V1/V2 loop of gp120 (S128N) that could partially restore the infectivity of mutant N260Q gp120 virus. However, the mutation S128N did not enhance any of the above-mentioned processes so its underlying compensatory mechanism must be a conformational effect that does not affect CD4 binding per se. Finally, we show that mutant N260Q gp160 was cleaved to gp120 and gp41 to a much lower extent than wild-type gp160, and that it was subject of lysosomal degradation to a higher extent than wild-type gp160 showing a prominent role of this process in the breakdown of N260-glycan-deleted gp160, which could not be counteracted by the S128N mutation. Moreover, at least part of the wild-type or mutant gp160 that is normally targeted for lysosomal degradation reached a conformation that enabled CD4 binding.
Conflict of interest statement
Figures



Similar articles
-
The disulfide loop of gp41 is critical to the furin recognition site of HIV gp160.Protein Sci. 2007 Jun;16(6):1236-41. doi: 10.1110/ps.072771407. Protein Sci. 2007. PMID: 17525470 Free PMC article.
-
Glycosylation of the core of the HIV-1 envelope subunit protein gp120 is not required for native trimer formation or viral infectivity.J Biol Chem. 2017 Jun 16;292(24):10197-10219. doi: 10.1074/jbc.M117.788919. Epub 2017 Apr 26. J Biol Chem. 2017. PMID: 28446609 Free PMC article.
-
Analysis of endoproteolytic cleavage and intracellular transport of human immunodeficiency virus type 1 envelope glycoproteins using mutant CD4 molecules bearing the transmembrane endoplasmic reticulum retention signal.J Gen Virol. 1993 Oct;74 ( Pt 10):2085-97. doi: 10.1099/0022-1317-74-10-2085. J Gen Virol. 1993. PMID: 8409933
-
Functional role of the glycan cluster of the human immunodeficiency virus type 1 transmembrane glycoprotein (gp41) ectodomain.J Virol. 1993 Jan;67(1):150-60. doi: 10.1128/JVI.67.1.150-160.1993. J Virol. 1993. PMID: 8093218 Free PMC article.
-
The highly conserved glycan at asparagine 260 of HIV-1 gp120 is indispensable for viral entry.J Biol Chem. 2011 Dec 16;286(50):42900-10. doi: 10.1074/jbc.M111.274456. Epub 2011 Oct 17. J Biol Chem. 2011. PMID: 22006924 Free PMC article.
Cited by
-
The HIV glycan shield as a target for broadly neutralizing antibodies.FEBS J. 2015 Dec;282(24):4679-91. doi: 10.1111/febs.13530. Epub 2015 Oct 23. FEBS J. 2015. PMID: 26411545 Free PMC article. Review.
-
Glycosylation States on Intact Proteins Determined by NMR Spectroscopy.Molecules. 2021 Jul 16;26(14):4308. doi: 10.3390/molecules26144308. Molecules. 2021. PMID: 34299586 Free PMC article.
-
Quantification of the Resilience and Vulnerability of HIV-1 Native Glycan Shield at Atomistic Detail.iScience. 2020 Nov 20;23(12):101836. doi: 10.1016/j.isci.2020.101836. eCollection 2020 Dec 18. iScience. 2020. PMID: 33319171 Free PMC article.
-
Several N-Glycans on the HIV Envelope Glycoprotein gp120 Preferentially Locate Near Disulphide Bridges and Are Required for Efficient Infectivity and Virus Transmission.PLoS One. 2015 Jun 29;10(6):e0130621. doi: 10.1371/journal.pone.0130621. eCollection 2015. PLoS One. 2015. PMID: 26121645 Free PMC article.
-
Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein.Cell Rep. 2016 Mar 22;14(11):2695-706. doi: 10.1016/j.celrep.2016.02.058. Epub 2016 Mar 10. Cell Rep. 2016. PMID: 26972002 Free PMC article.
References
-
- Reitter JN, Means RE, Desrosiers RC (1998) A role for carbohydrates in immune evasion in AIDS. Nat Med 4: 679–684. - PubMed
-
- Jitsuhara Y, Toyoda T, Itai T, Yamaguchi H (2002) Chaperone-like functions of high-mannose type and complex-type N-glycans and their molecular basis. J Biochem 132: 803–811. - PubMed
-
- Geyer H, Holschbach C, Hunsmann G, Schneider J (1988) Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J Biol Chem 263: 11760–11767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

