MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress
- PMID: 24962998
- PMCID: PMC4144780
- DOI: 10.1093/jxb/eru270
MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress
Abstract
Methionine sulfoxide reductases (MSRs) catalyse the reduction of oxidized methionine residues, thereby protecting proteins against oxidative stress. Accordingly, MSRs have been associated with stress responses, disease, and senescence in a taxonomically diverse array of organisms. However, the cytosolic substrates of MSRs in plants remain largely unknown. Here, we used a proteomic analysis strategy to identify MSRB7 substrates. We showed that two glutathione transferases (GSTs), GSTF2 and GSTF3, had fewer oxidized methionine (MetO) residues in MSRB7-overexpressing Arabidopsis thaliana plants than in wild-type plants. Conversely, GSTF2 and GSTF3 were highly oxidized and unstable in MSRB7-knockdown plants. MSRB7 was able to restore the MetO-GSTF2M100/104 and MetO-GSTF3M100 residues produced during oxidative stress. Furthermore, both GSTs were specifically induced by the oxidative stress inducer, methyl viologen. Our results indicate that specific GSTs are substrates of MSRs, which together provide a major line of defence against oxidative stress in A. thaliana.
Keywords: A. thaliana; LC-MS/MS; glutathione transferase; methionine sulfoxide reductase B (MSRB).; methyl viologen; oxidative stress.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures
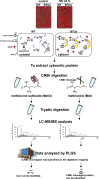

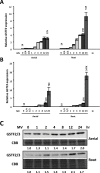

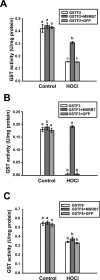
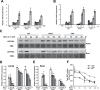
Similar articles
-
Arabidopsis root-abundant cytosolic methionine sulfoxide reductase B genes MsrB7 and MsrB8 are involved in tolerance to oxidative stress.Plant Cell Physiol. 2012 Oct;53(10):1707-19. doi: 10.1093/pcp/pcs114. Epub 2012 Aug 9. Plant Cell Physiol. 2012. PMID: 22885616
-
Arabidopsis thaliana methionine sulfoxide reductase B8 influences stress-induced cell death and effector-triggered immunity.Plant Mol Biol. 2017 Jan;93(1-2):109-120. doi: 10.1007/s11103-016-0550-z. Epub 2016 Nov 29. Plant Mol Biol. 2017. PMID: 27900506
-
Affinity chromatography: a valuable strategy to isolate substrates of methionine sulfoxide reductases?Antioxid Redox Signal. 2012 Jan 1;16(1):79-84. doi: 10.1089/ars.2011.4153. Epub 2011 Sep 1. Antioxid Redox Signal. 2012. PMID: 21882992
-
Recharging oxidative protein repair: catalysis by methionine sulfoxide reductases towards their amino acid, protein, and model substrates.Biochemistry (Mosc). 2012 Oct;77(10):1097-107. doi: 10.1134/S0006297912100021. Biochemistry (Mosc). 2012. PMID: 23157290 Review.
-
Roles of methionine suldfoxide reductases in antioxidant defense, protein regulation and survival.Curr Pharm Des. 2005;11(11):1451-7. doi: 10.2174/1381612053507846. Curr Pharm Des. 2005. PMID: 15853675 Review.
Cited by
-
MicroRNAs As Potential Targets for Abiotic Stress Tolerance in Plants.Front Plant Sci. 2016 Jun 14;7:817. doi: 10.3389/fpls.2016.00817. eCollection 2016. Front Plant Sci. 2016. PMID: 27379117 Free PMC article. Review.
-
(De)Activation (Ir)Reversibly or Degradation: Dynamics of Post-Translational Protein Modifications in Plants.Life (Basel). 2022 Feb 21;12(2):324. doi: 10.3390/life12020324. Life (Basel). 2022. PMID: 35207610 Free PMC article. Review.
-
Physiological Roles of Plant Methionine Sulfoxide Reductases in Redox Homeostasis and Signaling.Antioxidants (Basel). 2018 Aug 29;7(9):114. doi: 10.3390/antiox7090114. Antioxidants (Basel). 2018. PMID: 30158486 Free PMC article. Review.
-
Response of Multiple Tissues to Drought Revealed by a Weighted Gene Co-Expression Network Analysis in Foxtail Millet [Setaria italica (L.) P. Beauv.].Front Plant Sci. 2022 Jan 12;12:746166. doi: 10.3389/fpls.2021.746166. eCollection 2021. Front Plant Sci. 2022. PMID: 35095942 Free PMC article.
-
Spider mite egg extract modifies Arabidopsis response to future infestations.Sci Rep. 2021 Sep 6;11(1):17692. doi: 10.1038/s41598-021-97245-z. Sci Rep. 2021. PMID: 34489518 Free PMC article.
References
-
- Chen CJ, Tseng MC, Lin HJ, Lin TW, Chen YR. 2010. Visual indicator for surfactant abundance in MS-based membrane and general proteomics applications. Analytical Chemistry 82, 8283–8290 - PubMed
-
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of A. thaliana thaliana. The Plant Journal 16, 735–743 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

