Autophagic-lysosomal inhibition compromises ubiquitin-proteasome system performance in a p62 dependent manner in cardiomyocytes
- PMID: 24959866
- PMCID: PMC4069113
- DOI: 10.1371/journal.pone.0100715
Autophagic-lysosomal inhibition compromises ubiquitin-proteasome system performance in a p62 dependent manner in cardiomyocytes
Abstract
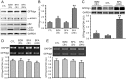
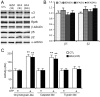
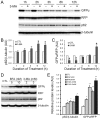
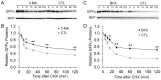
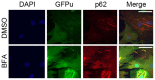
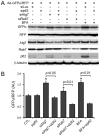
Intracellular protein degradation is primarily performed by the ubiquitin-proteasome system (UPS) and the autophagic-lysosomal pathway (ALP). The interplay between these two pathways has been rarely examined in intact animals and the mechanism underlying the interplay remains unclear. Hence, we sought to test in vivo and in vitro the impact of inhibition of the ALP on UPS proteolytic performance in cardiomyocytes and to explore the underlying mechanism. Transgenic mice ubiquitously expressing a surrogate UPS substrate (GFPdgn) were treated with bafilomycin-A1 (BFA) to inhibit the ALP. Myocardial and renal GFPdgn protein levels but not mRNA levels were increased at 24 hours but not 3 hours after the first injection of BFA. Myocardial protein abundance of key proteasome subunits and the activities of proteasomal peptidases were not discernibly altered by the treatment. In cultured neonatal rat ventricular myocytes (NRVMs), the surrogate UPS substrate GFPu and a control red fluorescence protein (RFP) were co-expressed to probe UPS performance. At 12 hours or 24 hours after ALP inhibition by 3-methyladenine (3-MA) or BFA, GFPu/RFP protein ratios and the protein half-life of GFPu were significantly increased, which is accompanied by increases in p62 proteins. Similar findings were obtained when ALP was inhibited genetically via silencing Atg7 or Rab7. ALP inhibition-induced increases in GFPu and p62 are co-localized in NRVMs. siRNA-mediated p62 knockdown prevented ALP inhibition from inducing GFPu accumulation in NRVMs. We conclude that in a p62-dependent fashion, ALP inhibition impairs cardiac UPS proteolytic performance in cardiomyocytes in vitro and in vivo.
Conflict of interest statement
Figures

Similar articles
-
The Calcineurin-TFEB-p62 Pathway Mediates the Activation of Cardiac Macroautophagy by Proteasomal Malfunction.Circ Res. 2020 Jul 31;127(4):502-518. doi: 10.1161/CIRCRESAHA.119.316007. Epub 2020 May 5. Circ Res. 2020. PMID: 32366200 Free PMC article.
-
A high-throughput screening identifies ZNF418 as a novel regulator of the ubiquitin-proteasome system and autophagy-lysosomal pathway.Autophagy. 2021 Oct;17(10):3124-3139. doi: 10.1080/15548627.2020.1856493. Epub 2020 Dec 27. Autophagy. 2021. PMID: 33249983 Free PMC article.
-
Tissue-Specific Impact of Autophagy Genes on the Ubiquitin-Proteasome System in C. elegans.Cells. 2020 Aug 8;9(8):1858. doi: 10.3390/cells9081858. Cells. 2020. PMID: 32784405 Free PMC article.
-
The ubiquitin-proteasome system (UPS) and the mechanism of action of bortezomib.Curr Pharm Des. 2011;17(15):1483-99. doi: 10.2174/138161211796197124. Curr Pharm Des. 2011. PMID: 21504411 Review.
-
The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity.Biochim Biophys Acta. 2015 Feb;1852(2):188-94. doi: 10.1016/j.bbadis.2014.07.028. Epub 2014 Aug 1. Biochim Biophys Acta. 2015. PMID: 25092168 Free PMC article. Review.
Cited by
-
cPKCγ-Modulated Autophagy in Neurons Alleviates Ischemic Injury in Brain of Mice with Ischemic Stroke Through Akt-mTOR Pathway.Transl Stroke Res. 2016 Dec;7(6):497-511. doi: 10.1007/s12975-016-0484-4. Epub 2016 Aug 10. Transl Stroke Res. 2016. PMID: 27510769
-
Systemic inhibition of neddylation by 3-day MLN4924 treatment regime does not impair autophagic flux in mouse hearts and brains.Am J Cardiovasc Dis. 2017 Dec 20;7(6):134-150. eCollection 2017. Am J Cardiovasc Dis. 2017. PMID: 29348974 Free PMC article.
-
Autophagy modulation: a potential therapeutic approach in cardiac hypertrophy.Am J Physiol Heart Circ Physiol. 2017 Aug 1;313(2):H304-H319. doi: 10.1152/ajpheart.00145.2017. Epub 2017 Jun 2. Am J Physiol Heart Circ Physiol. 2017. PMID: 28576834 Free PMC article. Review.
-
CYLD exaggerates pressure overload-induced cardiomyopathy via suppressing autolysosome efflux in cardiomyocytes.J Mol Cell Cardiol. 2020 Aug;145:59-73. doi: 10.1016/j.yjmcc.2020.06.004. Epub 2020 Jun 14. J Mol Cell Cardiol. 2020. PMID: 32553594 Free PMC article.
-
TLR-mediated aggresome-like induced structures comprise antimicrobial peptides and attenuate intracellular bacterial survival.Mol Biol Cell. 2024 Mar 1;35(3):ar34. doi: 10.1091/mbc.E23-09-0347. Epub 2024 Jan 3. Mol Biol Cell. 2024. PMID: 38170582 Free PMC article.
References
-
- Schlossarek S, Englmann DR, Sultan KR, Sauer M, Eschenhagen T, et al... (2012) Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic Res Cardiol 107: doi: 10.1007/s00395-00011-00235-00393 - PubMed
-
- Schlossarek S, Carrier L (2011) The ubiquitin-proteasome system in cardiomyopathies. Curr Opin Cardiol 26: 190–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

