Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics
- PMID: 24958771
- PMCID: PMC4068136
- DOI: 10.1083/jcb.201401126
Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics
Abstract
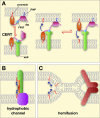
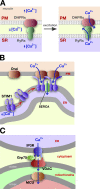
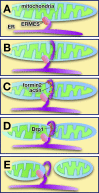
Regions of close apposition between two organelles, often referred to as membrane contact sites (MCSs), mostly form between the endoplasmic reticulum and a second organelle, although contacts between mitochondria and other organelles have also begun to be characterized. Although these contact sites have been noted since cells first began to be visualized with electron microscopy, the functions of most of these domains long remained unclear. The last few years have witnessed a dramatic increase in our understanding of MCSs, revealing the critical roles they play in intracellular signaling, metabolism, the trafficking of metabolites, and organelle inheritance, division, and transport.
Figures
Similar articles
-
Lipid transfer and signaling at organelle contact sites: the tip of the iceberg.Curr Opin Cell Biol. 2011 Aug;23(4):458-63. doi: 10.1016/j.ceb.2011.04.006. Curr Opin Cell Biol. 2011. PMID: 21555211 Free PMC article. Review.
-
Interacting organelles.Curr Opin Cell Biol. 2018 Aug;53:84-91. doi: 10.1016/j.ceb.2018.06.003. Epub 2018 Jul 2. Curr Opin Cell Biol. 2018. PMID: 30006038 Free PMC article. Review.
-
Organelle remodeling at membrane contact sites.J Struct Biol. 2016 Oct;196(1):15-19. doi: 10.1016/j.jsb.2016.05.003. Epub 2016 May 13. J Struct Biol. 2016. PMID: 27181417 Free PMC article. Review.
-
Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions.Trends Cell Biol. 2004 Sep;14(9):483-90. doi: 10.1016/j.tcb.2004.07.017. Trends Cell Biol. 2004. PMID: 15350976
-
Membrane contact sites, gateways for lipid homeostasis.Curr Opin Cell Biol. 2015 Apr;33:82-87. doi: 10.1016/j.ceb.2014.12.004. Epub 2015 Jan 6. Curr Opin Cell Biol. 2015. PMID: 25569848 Free PMC article. Review.
Cited by
-
Uptake and Metabolic Conversion of Exogenous Phosphatidylcholines Depending on Their Acyl Chain Structure in Arabidopsis thaliana.Int J Mol Sci. 2023 Dec 20;25(1):89. doi: 10.3390/ijms25010089. Int J Mol Sci. 2023. PMID: 38203257 Free PMC article.
-
Lipid Metabolism in Macrophages: Focus on Atherosclerosis.Biomedicines. 2020 Aug 1;8(8):262. doi: 10.3390/biomedicines8080262. Biomedicines. 2020. PMID: 32752275 Free PMC article. Review.
-
Dynamics and functions of lipid droplets.Nat Rev Mol Cell Biol. 2019 Mar;20(3):137-155. doi: 10.1038/s41580-018-0085-z. Nat Rev Mol Cell Biol. 2019. PMID: 30523332 Free PMC article. Review.
-
Ca2+ Dyshomeostasis Links Risk Factors to Neurodegeneration in Parkinson's Disease.Front Cell Neurosci. 2022 Apr 14;16:867385. doi: 10.3389/fncel.2022.867385. eCollection 2022. Front Cell Neurosci. 2022. PMID: 35496903 Free PMC article. Review.
-
The Expanding and Unexpected Functions of Mitochondria Contact Sites.Trends Cell Biol. 2019 Jul;29(7):580-590. doi: 10.1016/j.tcb.2019.02.009. Epub 2019 Mar 28. Trends Cell Biol. 2019. PMID: 30929794 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

