The pyrE Gene as a Bidirectional Selection Marker in Bifidobacterium Longum 105-A
- PMID: 24936363
- PMCID: PMC4034322
- DOI: 10.12938/bmfh.32.59
The pyrE Gene as a Bidirectional Selection Marker in Bifidobacterium Longum 105-A
Abstract
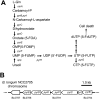
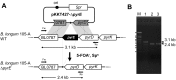
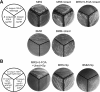
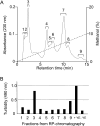
We constructed a deletion mutant of the pyrE gene in Bifidobacterium longum 105-A. A pyrE knockout cassette was cloned into pKKT427, a Bifidobacterium-Escherichia coli shuttle vector, and then introduced into B. longum 105-A by electroporation. The transformants were propagated and spread onto MRS plates containing 5-fluoroorotic acid (5-FOA) and uracil. 5-FOA-resistant mutants were obtained at a frequency of 4.7 × 10(-5) integrations per cell. To perform pyrE gene complementation, the pyrE gene was amplified by PCR and used to construct a complementation plasmid (pKKT427-pyrE (+)). B. longum 105-A ∆pyrE harboring this plasmid could not grow on MRS plates containing 5-FOA, uracil and spectinomycin. We also developed a chemically defined medium (bifidobacterial minimal medium; BMM) containing inorganic salts, glucose, vitamins, isoleucine and tyrosine for positive selection of pyrE transformants. B. longum 105-A ∆pyrE could not grow on BMM agar, but the same strain harboring pKKT427-pyrE (+) could. Thus, pyrE can be used as a counterselection marker in B. longum 105-A and potentially other Bifidobacterium species as well. We demonstrated the effectiveness of this system by constructing a knockout mutant of the xynF gene in B. longum 105-A by using the pyrE gene as a counterselection marker. This pyrE-based selection system will contribute to genetic studies of bifidobacteria.
Keywords: 5-FOA; B. longum 105-A; bifidobacterial minimal medium (BMM); gene inactivation; gene knockout; homologous recombination; pyrimidine metabolism.
Figures
Similar articles
-
A targeted gene knockout method using a newly constructed temperature-sensitive plasmid mediated homologous recombination in Bifidobacterium longum.Appl Microbiol Biotechnol. 2012 Jul;95(2):499-509. doi: 10.1007/s00253-012-4090-4. Epub 2012 May 27. Appl Microbiol Biotechnol. 2012. PMID: 22639142
-
Development of a pyrE-based selective system for Thermotoga sp. strain RQ7.Extremophiles. 2017 Mar;21(2):297-306. doi: 10.1007/s00792-016-0902-2. Epub 2016 Dec 7. Extremophiles. 2017. PMID: 27928679
-
Transformation of Bifidobacterium longum with pRM2, a constructed Escherichia coli-B. longum shuttle vector.Plasmid. 1994 Sep;32(2):208-11. doi: 10.1006/plas.1994.1056. Plasmid. 1994. PMID: 7846144
-
A roadmap for gene system development in Clostridium.Anaerobe. 2016 Oct;41:104-112. doi: 10.1016/j.anaerobe.2016.05.011. Epub 2016 May 24. Anaerobe. 2016. PMID: 27234263 Free PMC article. Review.
-
Technological advances in bifidobacterial molecular genetics: application to functional genomics and medical treatments.Biosci Microbiota Food Health. 2012;31(2):15-25. doi: 10.12938/bmfh.31.15. Epub 2012 Apr 20. Biosci Microbiota Food Health. 2012. PMID: 24936345 Free PMC article. Review.
Cited by
-
Genetic tools for the electrotroph Sporomusa ovata and autotrophic biosynthesis.Appl Environ Microbiol. 2024 Jan 24;90(1):e0175723. doi: 10.1128/aem.01757-23. Epub 2023 Dec 20. Appl Environ Microbiol. 2024. PMID: 38117058 Free PMC article.
-
A novel SfaNI-like restriction-modification system in Caldicellulosiruptor extents the genetic engineering toolbox for this genus.PLoS One. 2022 Dec 29;17(12):e0279562. doi: 10.1371/journal.pone.0279562. eCollection 2022. PLoS One. 2022. PMID: 36580476 Free PMC article.
-
Bacterial Genetic Engineering by Means of Recombineering for Reverse Genetics.Front Microbiol. 2020 Sep 11;11:548410. doi: 10.3389/fmicb.2020.548410. eCollection 2020. Front Microbiol. 2020. PMID: 33013782 Free PMC article. Review.
-
Identifying the essential nutritional requirements of the probiotic bacteria Bifidobacterium animalis and Bifidobacterium longum through genome-scale modeling.NPJ Syst Biol Appl. 2021 Dec 9;7(1):47. doi: 10.1038/s41540-021-00207-4. NPJ Syst Biol Appl. 2021. PMID: 34887435 Free PMC article.
-
Methionine utilization by bifidobacteria: possible existence of a reverse transsulfuration pathway.Biosci Microbiota Food Health. 2021;40(1):80-83. doi: 10.12938/bmfh.2020-031. Epub 2020 Aug 22. Biosci Microbiota Food Health. 2021. PMID: 33520573 Free PMC article.
References
-
- Stackebrandt E, Rainey FA, WardRainey NL. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 47: 479–491
-
- Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B, O’Sullivan DJ. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9: 247 - PMC - PubMed
-
- Yasui K, Tabata M, Yamada S, Abe T, Ikemura T, Osawa R, Suzuki T. 2009. Intra-species diversity between seven Bifidobacterium adolescentis strains identified by genome-wide tiling array analysis. Biosci Biotechnol Biochem 73: 1422–1424 - PubMed
-
- Suzuki T, Yasui K. 2011. Strain engineering: Methods and protocols, methods in molecular biology, In Williams, J. A. (ed.), Chapter 18, pp. 309–326, Humana Press, New York.
LinkOut - more resources
Full Text Sources
Other Literature Sources
