CD4-induced activation in a soluble HIV-1 Env trimer
- PMID: 24931470
- PMCID: PMC4231881
- DOI: 10.1016/j.str.2014.05.001
CD4-induced activation in a soluble HIV-1 Env trimer
Abstract
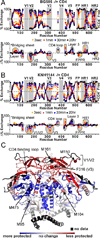
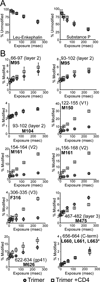
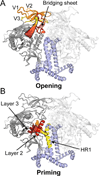
The HIV envelope glycoprotein (Env) trimer undergoes receptor-induced conformational changes that drive fusion of the viral and cellular membranes. Env conformational changes have been observed using low-resolution electron microscopy, but only large-scale rearrangements have been visible. Here, we use hydrogen-deuterium exchange and oxidative labeling to gain a more precise understanding of the unliganded and CD4-bound forms of soluble Env trimers (SOSIP.664), including their glycan composition. CD4 activation induces the reorganization of bridging sheet elements, V1/V2 and V3, much of the gp120 inner domain, and the gp41 fusion subunit. Two CD4 binding site-targeted inhibitors have substantially different effects: NBD-556 partially mimics CD4-induced destabilization of the V1/V2 and V3 crown, whereas BMS-806 only affects regions around the gp120/gp41 interface. The structural information presented here increases our knowledge of CD4- and small molecule-induced conformational changes in Env and the allosteric pathways that lead to membrane fusion.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures




Comment in
-
The HIV-1 Env trimer in HD.Structure. 2014 Jul 8;22(7):935-6. doi: 10.1016/j.str.2014.06.004. Structure. 2014. PMID: 25007222 Free PMC article.
Similar articles
-
Characterization of Human Immunodeficiency Virus (HIV-1) Envelope Glycoprotein Variants Selected for Resistance to a CD4-Mimetic Compound.J Virol. 2022 Sep 14;96(17):e0063622. doi: 10.1128/jvi.00636-22. Epub 2022 Aug 18. J Virol. 2022. PMID: 35980207 Free PMC article.
-
Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):E7151-E7158. doi: 10.1073/pnas.1615939113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799557 Free PMC article.
-
Conformational Differences between Functional Human Immunodeficiency Virus Envelope Glycoprotein Trimers and Stabilized Soluble Trimers.J Virol. 2019 Jan 17;93(3):e01709-18. doi: 10.1128/JVI.01709-18. Print 2019 Feb 1. J Virol. 2019. PMID: 30429345 Free PMC article.
-
Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer.PLoS One. 2015 Apr 7;10(4):e0122111. doi: 10.1371/journal.pone.0122111. eCollection 2015. PLoS One. 2015. PMID: 25849367 Free PMC article.
-
Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
Cited by
-
Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env.Nat Struct Mol Biol. 2015 Jul;22(7):522-31. doi: 10.1038/nsmb.3051. Epub 2015 Jun 22. Nat Struct Mol Biol. 2015. PMID: 26098315 Free PMC article.
-
Oriented display of HIV-1 Env trimers by a novel coupling strategy enhances B cell activation and phagocytosis.Front Immunol. 2024 Feb 8;15:1344346. doi: 10.3389/fimmu.2024.1344346. eCollection 2024. Front Immunol. 2024. PMID: 38390320 Free PMC article.
-
Exploring the potential of monoclonal antibody therapeutics for HIV-1 eradication.AIDS Res Hum Retroviruses. 2015 Jan;31(1):13-24. doi: 10.1089/aid.2014.0235. AIDS Res Hum Retroviruses. 2015. PMID: 25385703 Free PMC article.
-
Structure and immune recognition of trimeric pre-fusion HIV-1 Env.Nature. 2014 Oct 23;514(7523):455-61. doi: 10.1038/nature13808. Epub 2014 Oct 8. Nature. 2014. PMID: 25296255 Free PMC article.
-
Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer.Science. 2016 Mar 4;351(6277):1043-8. doi: 10.1126/science.aad2450. Science. 2016. PMID: 26941313 Free PMC article.
References
-
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. - PMC - PubMed
-
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI027767/AI/NIAID NIH HHS/United States
- R01-GM099989/GM/NIGMS NIH HHS/United States
- UM1 AI100663/AI/NIAID NIH HHS/United States
- R00 GM080352/GM/NIGMS NIH HHS/United States
- R37-AI36082/AI/NIAID NIH HHS/United States
- R01-AI084817/AI/NIAID NIH HHS/United States
- R01 AI084817/AI/NIAID NIH HHS/United States
- R01 GM099989/GM/NIGMS NIH HHS/United States
- R56 AI084817/AI/NIAID NIH HHS/United States
- R37 AI036082/AI/NIAID NIH HHS/United States
- P41-GM103393/GM/NIGMS NIH HHS/United States
- T32-GM007750/GM/NIGMS NIH HHS/United States
- P01-AI82362/AI/NIAID NIH HHS/United States
- R01-AI41420/AI/NIAID NIH HHS/United States
- 280829/ERC_/European Research Council/International
- F32 GM097805/GM/NIGMS NIH HHS/United States
- R00-GM080352/GM/NIGMS NIH HHS/United States
- F32-GM097805/GM/NIGMS NIH HHS/United States
- P41 RR001209/RR/NCRR NIH HHS/United States
- T32 GM007750/GM/NIGMS NIH HHS/United States
- P41-RR001209/RR/NCRR NIH HHS/United States
- P30-AI027767/AI/NIAID NIH HHS/United States
- R01 AI041420/AI/NIAID NIH HHS/United States
- P01 AI082362/AI/NIAID NIH HHS/United States
- P41 GM103393/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

