Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53
- PMID: 24919155
- PMCID: PMC4112218
- DOI: 10.1038/nature13298
Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53
Abstract
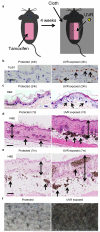
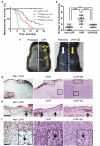
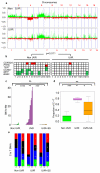
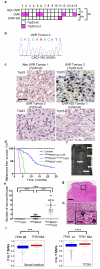
Cutaneous melanoma is epidemiologically linked to ultraviolet radiation (UVR), but the molecular mechanisms by which UVR drives melanomagenesis remain unclear. The most common somatic mutation in melanoma is a V600E substitution in BRAF, which is an early event. To investigate how UVR accelerates oncogenic BRAF-driven melanomagenesis, we used a BRAF(V600E) mouse model. In mice expressing BRAF(V600E) in their melanocytes, a single dose of UVR that mimicked mild sunburn in humans induced clonal expansion of the melanocytes, and repeated doses of UVR increased melanoma burden. Here we show that sunscreen (UVA superior, UVB sun protection factor (SPF) 50) delayed the onset of UVR-driven melanoma, but only provided partial protection. The UVR-exposed tumours showed increased numbers of single nucleotide variants and we observed mutations (H39Y, S124F, R245C, R270C, C272G) in the Trp53 tumour suppressor in approximately 40% of cases. TP53 is an accepted UVR target in human non-melanoma skin cancer, but is not thought to have a major role in melanoma. However, we show that, in mice, mutant Trp53 accelerated BRAF(V600E)-driven melanomagenesis, and that TP53 mutations are linked to evidence of UVR-induced DNA damage in human melanoma. Thus, we provide mechanistic insight into epidemiological data linking UVR to acquired naevi in humans. Furthermore, we identify TP53/Trp53 as a UVR-target gene that cooperates with BRAF(V600E) to induce melanoma, providing molecular insight into how UVR accelerates melanomagenesis. Our study validates public health campaigns that promote sunscreen protection for individuals at risk of melanoma.
Figures
Comment in
-
UV and melanoma: the TP53 link.Cell Res. 2014 Oct;24(10):1157-8. doi: 10.1038/cr.2014.95. Epub 2014 Jul 18. Cell Res. 2014. PMID: 25033756 Free PMC article.
-
TP53 in the UV spotlight: a bona fide driver of melanoma.Pigment Cell Melanoma Res. 2014 Nov;27(6):1010-1. doi: 10.1111/pcmr.12302. Epub 2014 Sep 2. Pigment Cell Melanoma Res. 2014. PMID: 25102761 No abstract available.
Similar articles
-
UV and melanoma: the TP53 link.Cell Res. 2014 Oct;24(10):1157-8. doi: 10.1038/cr.2014.95. Epub 2014 Jul 18. Cell Res. 2014. PMID: 25033756 Free PMC article.
-
Melanocytic nevus-like hyperplasia and melanoma in transgenic BRAFV600E mice.Oncogene. 2009 Jun 11;28(23):2289-98. doi: 10.1038/onc.2009.95. Epub 2009 Apr 27. Oncogene. 2009. PMID: 19398955 Free PMC article.
-
Telomere dysfunction in Tert knockout mice delays BrafV600E -induced melanoma development.Int J Cancer. 2024 Feb 1;154(3):548-560. doi: 10.1002/ijc.34713. Epub 2023 Sep 20. Int J Cancer. 2024. PMID: 37727982 Free PMC article.
-
Macroenvironment-gene-microenvironment interactions in ultraviolet radiation-induced melanomagenesis.Adv Cancer Res. 2019;144:1-54. doi: 10.1016/bs.acr.2019.03.008. Epub 2019 Apr 23. Adv Cancer Res. 2019. PMID: 31349897 Review.
-
UV-Induced Molecular Signaling Differences in Melanoma and Non-melanoma Skin Cancer.Adv Exp Med Biol. 2017;996:27-40. doi: 10.1007/978-3-319-56017-5_3. Adv Exp Med Biol. 2017. PMID: 29124688 Review.
Cited by
-
From Proteomic Mapping to Invasion-Metastasis-Cascade Systemic Biomarkering and Targeted Drugging of Mutant BRAF-Dependent Human Cutaneous Melanomagenesis.Cancers (Basel). 2021 Apr 22;13(9):2024. doi: 10.3390/cancers13092024. Cancers (Basel). 2021. PMID: 33922182 Free PMC article.
-
Toward predicting metastatic progression of melanoma based on gene expression data.Pigment Cell Melanoma Res. 2015 Jul;28(4):453-63. doi: 10.1111/pcmr.12374. Epub 2015 Apr 24. Pigment Cell Melanoma Res. 2015. PMID: 25847062 Free PMC article.
-
UVA Radiation, DNA Damage, and Melanoma.ACS Omega. 2022 Sep 8;7(37):32936-32948. doi: 10.1021/acsomega.2c04424. eCollection 2022 Sep 20. ACS Omega. 2022. PMID: 36157735 Free PMC article. Review.
-
Oncogenic BRAF(V600E) Induces Clastogenesis and UVB Hypersensitivity.Cancers (Basel). 2015 Jun 17;7(2):1072-90. doi: 10.3390/cancers7020825. Cancers (Basel). 2015. PMID: 26091525 Free PMC article.
-
Melanocyte stem cells in the skin: Origin, biological characteristics, homeostatic maintenance and therapeutic potential.Clin Transl Med. 2024 May;14(5):e1720. doi: 10.1002/ctm2.1720. Clin Transl Med. 2024. PMID: 38778457 Free PMC article. Review.
References
-
- Whiteman DC, et al. Melanocytic nevi, solar keratoses, and divergent pathways to cutaneous melanoma. J Natl Cancer Inst. 2003;95:806–812. - PubMed
-
- Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N Engl J Med. 1999;340:1341–1348. doi:10.1056/NEJM199904293401707. - PubMed
-
- Harrison SL, MacLennan R, Speare R, Wronski I. Sun exposure and melanocytic naevi in young Australian children. Lancet. 1994;344:1529–1532. - PubMed
METHODS REFERENCES
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

