Quantitative temporal viromics: an approach to investigate host-pathogen interaction
- PMID: 24906157
- PMCID: PMC4048463
- DOI: 10.1016/j.cell.2014.04.028
Quantitative temporal viromics: an approach to investigate host-pathogen interaction
Abstract
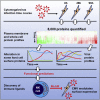
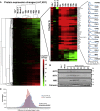
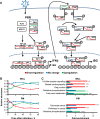
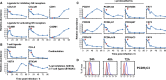
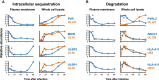
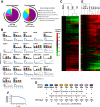
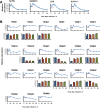
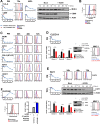
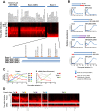
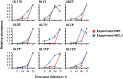
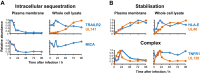
A systematic quantitative analysis of temporal changes in host and viral proteins throughout the course of a productive infection could provide dynamic insights into virus-host interaction. We developed a proteomic technique called "quantitative temporal viromics" (QTV), which employs multiplexed tandem-mass-tag-based mass spectrometry. Human cytomegalovirus (HCMV) is not only an important pathogen but a paradigm of viral immune evasion. QTV detailed how HCMV orchestrates the expression of >8,000 cellular proteins, including 1,200 cell-surface proteins to manipulate signaling pathways and counterintrinsic, innate, and adaptive immune defenses. QTV predicted natural killer and T cell ligands, as well as 29 viral proteins present at the cell surface, potential therapeutic targets. Temporal profiles of >80% of HCMV canonical genes and 14 noncanonical HCMV open reading frames were defined. QTV is a powerful method that can yield important insights into viral infection and is applicable to any virus with a robust in vitro model.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
High-Definition Analysis of Host Protein Stability during Human Cytomegalovirus Infection Reveals Antiviral Factors and Viral Evasion Mechanisms.Cell Host Microbe. 2018 Sep 12;24(3):447-460.e11. doi: 10.1016/j.chom.2018.07.011. Epub 2018 Aug 16. Cell Host Microbe. 2018. PMID: 30122656 Free PMC article.
-
Dendritic cells in cytomegalovirus infection: viral evasion and host countermeasures.APMIS. 2009 May;117(5-6):413-26. doi: 10.1111/j.1600-0463.2009.02449.x. APMIS. 2009. PMID: 19400865 Review.
-
Genetic Variability of Human Cytomegalovirus Clinical Isolates Correlates With Altered Expression of Natural Killer Cell-Activating Ligands and IFN-γ.Front Immunol. 2021 Apr 9;12:532484. doi: 10.3389/fimmu.2021.532484. eCollection 2021. Front Immunol. 2021. PMID: 33897679 Free PMC article.
-
The tiers and dimensions of evasion of the type I interferon response by human cytomegalovirus.J Mol Biol. 2013 Dec 13;425(24):4857-71. doi: 10.1016/j.jmb.2013.08.023. Epub 2013 Sep 5. J Mol Biol. 2013. PMID: 24013068 Free PMC article. Review.
-
Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection.Nat Commun. 2020 Feb 10;11(1):806. doi: 10.1038/s41467-020-14586-5. Nat Commun. 2020. PMID: 32041945 Free PMC article.
Cited by
-
A conditionally replication-defective cytomegalovirus vaccine elicits potent and diverse functional monoclonal antibodies in a phase I clinical trial.NPJ Vaccines. 2021 Jun 2;6(1):79. doi: 10.1038/s41541-021-00342-3. NPJ Vaccines. 2021. PMID: 34078915 Free PMC article.
-
The Cellular Proteins Grb2 and DDX3 Are Increased upon Human Cytomegalovirus Infection and Act in a Proviral Fashion.PLoS One. 2015 Jun 29;10(6):e0131614. doi: 10.1371/journal.pone.0131614. eCollection 2015. PLoS One. 2015. PMID: 26121620 Free PMC article.
-
Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS.Nature. 2015 Jul 9;523(7559):183-188. doi: 10.1038/nature14610. Epub 2015 Jul 1. Nature. 2015. PMID: 26131937 Free PMC article.
-
Cytomegalovirus immune evasion of myeloid lineage cells.Med Microbiol Immunol. 2015 Jun;204(3):367-82. doi: 10.1007/s00430-015-0403-4. Epub 2015 Mar 17. Med Microbiol Immunol. 2015. PMID: 25776081 Review.
-
Studies on the Contribution of Human Cytomegalovirus UL21a and UL97 to Viral Growth and Inactivation of the Anaphase-Promoting Complex/Cyclosome (APC/C) E3 Ubiquitin Ligase Reveal a Unique Cellular Mechanism for Downmodulation of the APC/C Subunits APC1, APC4, and APC5.J Virol. 2015 Jul;89(13):6928-39. doi: 10.1128/JVI.00403-15. Epub 2015 Apr 22. J Virol. 2015. PMID: 25903336 Free PMC article.
References
-
- Alexander B.T., Hladnik L.M., Augustin K.M., Casabar E., McKinnon P.S., Reichley R.M., Ritchie D.J., Westervelt P., Dubberke E.R. Use of cytomegalovirus intravenous immune globulin for the adjunctive treatment of cytomegalovirus in hematopoietic stem cell transplant recipients. Pharmacotherapy. 2010;30:554–561. - PMC - PubMed
-
- Bonneville M., O’Brien R.L., Born W.K. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010;10:467–478. - PubMed
Supplemental References
-
- Ahn K., Gruhler A., Galocha B., Jones T.R., Wiertz E.J., Ploegh H.L., Peterson P.A., Yang Y., Früh K. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity. 1997;6:613–621. - PubMed
-
- Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13:11–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 101835/WT_/Wellcome Trust/United Kingdom
- WT090323MA/WT_/Wellcome Trust/United Kingdom
- G1000236/WT_/Wellcome Trust/United Kingdom
- G1000236/MRC_/Medical Research Council/United Kingdom
- R01 GM067945/GM/NIGMS NIH HHS/United States
- GM067945/GM/NIGMS NIH HHS/United States
- MR/L018373/1/MRC_/Medical Research Council/United Kingdom
- MR/L008734/1/MRC_/Medical Research Council/United Kingdom
- G0901119/MRC_/Medical Research Council/United Kingdom
- 093966/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- 100140/WT_/Wellcome Trust/United Kingdom
- 084957/Z/08/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

