Rab18 and a Rab18 GEF complex are required for normal ER structure
- PMID: 24891604
- PMCID: PMC4050724
- DOI: 10.1083/jcb.201403026
Rab18 and a Rab18 GEF complex are required for normal ER structure
Abstract
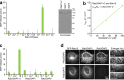
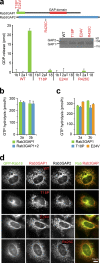
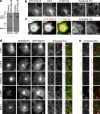
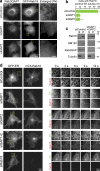
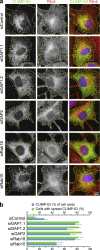
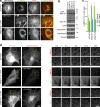
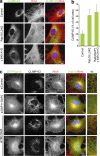
The ancestral Rab GTPase Rab18 and both subunits of the Rab3GAP complex are mutated in the human neurological and developmental disorder Warburg Micro syndrome. Here, we demonstrate that the Rab3GAP complex is a specific Rab18 guanine nucleotide exchange factor (GEF). The Rab3GAP complex localizes to the endoplasmic reticulum (ER) and is necessary for ER targeting of Rab18. It is also sufficient to promote membrane recruitment of Rab18. Disease-associated point mutations of conserved residues in either the Rab3GAP1 (T18P and E24V) or Rab3GAP2 (R426C) subunits result in loss of the Rab18 GEF and membrane-targeting activities. Supporting the view that Rab18 activity is important for ER structure, in the absence of either Rab3GAP subunit or Rab18 function, ER tubular networks marked by reticulon 4 were disrupted, and ER sheets defined by CLIMP-63 spread out into the cell periphery. Micro syndrome is therefore a disease characterized by direct loss of Rab18 function or loss of Rab18 activation at the ER by its GEF Rab3GAP.
© 2014 Gerondopoulos et al.
Figures
Similar articles
-
Warburg Micro syndrome is caused by RAB18 deficiency or dysregulation.Open Biol. 2015 Jun;5(6):150047. doi: 10.1098/rsob.150047. Open Biol. 2015. PMID: 26063829 Free PMC article.
-
Biochemical and structural characterization of Rab3GAP reveals insights into Rab18 nucleotide exchange activity.Nat Commun. 2025 Jan 8;16(1):479. doi: 10.1038/s41467-025-55828-8. Nat Commun. 2025. PMID: 39779760 Free PMC article.
-
The Warburg Micro Syndrome-associated Rab3GAP-Rab18 module promotes autolysosome maturation through the Vps34 Complex I.FEBS J. 2021 Jan;288(1):190-211. doi: 10.1111/febs.15313. Epub 2020 Apr 22. FEBS J. 2021. PMID: 32248620
-
RAB3GAP1, RAB3GAP2 and RAB18: disease genes in Micro and Martsolf syndromes.Biochem Soc Trans. 2012 Dec 1;40(6):1394-7. doi: 10.1042/BST20120169. Biochem Soc Trans. 2012. PMID: 23176487 Review.
-
Rab18: new insights into the function of an essential protein.Cell Mol Life Sci. 2019 May;76(10):1935-1945. doi: 10.1007/s00018-019-03050-3. Epub 2019 Mar 4. Cell Mol Life Sci. 2019. PMID: 30830238 Free PMC article. Review.
Cited by
-
Novel RAB3GAP1 Mutations Causing Warburg Micro Syndrome in Two Italian Sisters.J Pediatr Neurosci. 2017 Oct-Dec;12(4):360-362. doi: 10.4103/jpn.JPN_45_17. J Pediatr Neurosci. 2017. PMID: 29675078 Free PMC article.
-
A Homozygous RAB3GAP1:c.743delC Mutation in Rottweilers with Neuronal Vacuolation and Spinocerebellar Degeneration.J Vet Intern Med. 2016 May;30(3):813-8. doi: 10.1111/jvim.13921. Epub 2016 Mar 10. J Vet Intern Med. 2016. PMID: 26968732 Free PMC article.
-
ER residential chaperone GRP78 unconventionally relocalizes to the cell surface via endosomal transport.Cell Mol Life Sci. 2021 Jun;78(12):5179-5195. doi: 10.1007/s00018-021-03849-z. Epub 2021 May 11. Cell Mol Life Sci. 2021. PMID: 33974094 Free PMC article.
-
On-demand autophagic network adaptations upon limited lipid availability.Autophagy. 2020 Jul;16(7):1348-1350. doi: 10.1080/15548627.2020.1753926. Epub 2020 Apr 16. Autophagy. 2020. PMID: 32295467 Free PMC article.
-
Functional Genomics Identifies Tis21-Dependent Mechanisms and Putative Cancer Drug Targets Underlying Medulloblastoma Shh-Type Development.Front Pharmacol. 2016 Nov 30;7:449. doi: 10.3389/fphar.2016.00449. eCollection 2016. Front Pharmacol. 2016. PMID: 27965576 Free PMC article.
References
-
- Aligianis I.A., Morgan N.V., Mione M., Johnson C.A., Rosser E., Hennekam R.C., Adams G., Trembath R.C., Pilz D.T., Stoodley N., et al. 2006. Mutation in Rab3 GTPase-activating protein (RAB3GAP) noncatalytic subunit in a kindred with Martsolf syndrome. Am. J. Hum. Genet. 78:702–707 10.1086/502681 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

