LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint
- PMID: 24857549
- PMCID: PMC4103188
- DOI: 10.1016/j.molcel.2014.04.025
LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint
Abstract
The p53-regulated long noncoding RNA lincRNA-p21 has been proposed to act in trans via several mechanisms ranging from repressing genes in the p53 transcriptional network to regulating mRNA translation and protein stability. To further examine lincRNA-p21 function, we generated a conditional knockout mouse model. We find that lincRNA-p21 predominantly functions in cis to activate expression of its neighboring gene, p21. Mechanistically, we show that lincRNA-p21 acts in concert with hnRNP-K as a coactivator for p53-dependent p21 transcription. Additional phenotypes of lincRNA-p21 deficiency could be attributed to diminished p21 levels, including deregulated expression and altered chromatin state of some Polycomb target genes, a defective G1/S checkpoint, increased proliferation rates, and enhanced reprogramming efficiency. These findings indicate that lincRNA-p21 affects global gene expression and influences the p53 tumor suppressor pathway by acting in cis as a locus-restricted coactivator for p53-mediated p21 expression.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

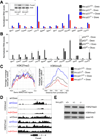
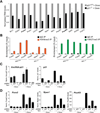

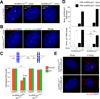
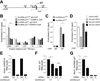
Similar articles
-
LincRNA-p21 acts as a mediator of ING1b-induced apoptosis.Cell Death Dis. 2015 Mar 5;6(3):e1668. doi: 10.1038/cddis.2015.15. Cell Death Dis. 2015. PMID: 25741593 Free PMC article.
-
Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes.Cell Death Dis. 2015 Mar 19;6(3):e1700. doi: 10.1038/cddis.2015.67. Cell Death Dis. 2015. PMID: 25789975 Free PMC article.
-
p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma.Mol Cancer. 2019 Mar 11;18(1):38. doi: 10.1186/s12943-019-0993-3. Mol Cancer. 2019. PMID: 30857539 Free PMC article.
-
LincRNa-p21: function and mechanism in cancer.Med Oncol. 2017 May;34(5):98. doi: 10.1007/s12032-017-0959-5. Epub 2017 Apr 19. Med Oncol. 2017. PMID: 28425074 Review.
-
The role of lincRNA-p21 in regulating the biology of cancer cells.Hum Cell. 2022 Nov;35(6):1640-1649. doi: 10.1007/s13577-022-00768-4. Epub 2022 Aug 15. Hum Cell. 2022. PMID: 35969349 Review.
Cited by
-
Targeting DNA Damage Response in the Radio(Chemo)therapy of Non-Small Cell Lung Cancer.Int J Mol Sci. 2016 May 31;17(6):839. doi: 10.3390/ijms17060839. Int J Mol Sci. 2016. PMID: 27258253 Free PMC article. Review.
-
Signaling by LncRNAs: Structure, Cellular Homeostasis, and Disease Pathology.Cells. 2022 Aug 13;11(16):2517. doi: 10.3390/cells11162517. Cells. 2022. PMID: 36010595 Free PMC article. Review.
-
Role of long non-coding RNA in drug resistance in non-small cell lung cancer.Thorac Cancer. 2018 Jul;9(7):761-768. doi: 10.1111/1759-7714.12652. Epub 2018 May 3. Thorac Cancer. 2018. PMID: 29726094 Free PMC article. Review.
-
Human Long Noncoding RNA Interactome: Detection, Characterization and Function.Int J Mol Sci. 2020 Feb 4;21(3):1027. doi: 10.3390/ijms21031027. Int J Mol Sci. 2020. PMID: 32033158 Free PMC article. Review.
-
Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer.Front Oncol. 2020 Mar 20;10:285. doi: 10.3389/fonc.2020.00285. eCollection 2020. Front Oncol. 2020. PMID: 32266130 Free PMC article. Review.
References
-
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377(6549):552–557. - PubMed
-
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82(4):675–684. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

