Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome
- PMID: 24837403
- PMCID: PMC7107391
- DOI: 10.1093/infdis/jiu292
Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome
Abstract
Background: Analysis of clinical samples from patients with new viral infections is critical to confirm the diagnosis, to specify the viral load, and to sequence data necessary for characterizing the viral kinetics, transmission, and evolution. We analyzed samples from 112 patients infected with the recently discovered Middle East respiratory syndrome coronavirus (MERS-CoV).
Methods: Respiratory tract samples from cases of MERS-CoV infection confirmed by polymerase chain reaction (PCR) were investigated to determine the MERS-CoV load and fraction of the MERS-CoV genome. These values were analyzed to determine associations with clinical sample type.
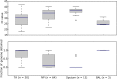
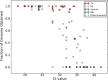
Results: Samples from 112 individuals in which MERS-CoV was detected by PCR were analyzed, of which 13 were sputum samples, 64 were nasopharyngeal swab specimens, 30 were tracheal aspirates, and 3 were bronchoalveolar lavage specimens; 2 samples were of unknown origin. Tracheal aspirates yielded significantly higher MERS-CoV loads, compared with nasopharyngeal swab specimens (P = .005) and sputum specimens (P = .0001). Tracheal aspirates had viral loads similar to those in bronchoalveolar lavage samples (P = .3079). Bronchoalveolar lavage samples and tracheal aspirates had significantly higher genome fraction than nasopharyngeal swab specimens (P = .0095 and P = .0002, respectively) and sputum samples (P = .0009 and P = .0001, respectively). The genome yield from tracheal aspirates and bronchoalveolar lavage samples were similar (P = .1174).
Conclusions: Lower respiratory tract samples yield significantly higher MERS-CoV loads and genome fractions than upper respiratory tract samples.
Keywords: Ct value; MERS-CoV; Middle East; RT-PCR; clinical; coronavirus; diagnosis; genome fraction; molecular; screening; viral load.
© The Author 2014. Published by Oxford University Press on behalf of the Infectious Diseases Society of America. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Replicative virus shedding in the respiratory tract of patients with Middle East respiratory syndrome coronavirus infection.Int J Infect Dis. 2018 Jul;72:8-10. doi: 10.1016/j.ijid.2018.05.003. Epub 2018 May 9. Int J Infect Dis. 2018. PMID: 29753119 Free PMC article.
-
Kinetics and pattern of viral excretion in biological specimens of two MERS-CoV cases.J Clin Virol. 2014 Oct;61(2):275-8. doi: 10.1016/j.jcv.2014.07.002. Epub 2014 Jul 12. J Clin Virol. 2014. PMID: 25073585 Free PMC article.
-
Middle East respiratory syndrome coronavirus (MERS-CoV) viral shedding in the respiratory tract: an observational analysis with infection control implications.Int J Infect Dis. 2014 Dec;29:307-8. doi: 10.1016/j.ijid.2014.10.002. Epub 2014 Oct 24. Int J Infect Dis. 2014. PMID: 25448335 Free PMC article.
-
Emerging Developments on Pathogenicity, Molecular Virulence, Epidemiology and Clinical Symptoms of Current Middle East Respiratory Syndrome Coronavirus (MERS-CoV).Hayati. 2017 Apr;24(2):53-56. doi: 10.1016/j.hjb.2017.08.001. Epub 2017 Sep 14. Hayati. 2017. PMID: 32288937 Free PMC article. Review.
-
Pathogenesis of Middle East respiratory syndrome coronavirus.J Pathol. 2015 Jan;235(2):175-84. doi: 10.1002/path.4458. J Pathol. 2015. PMID: 25294366 Free PMC article. Review.
Cited by
-
Temporal Profiles of Antibody Responses, Cytokines, and Survival of COVID-19 Patients: A Retrospective Cohort.Engineering (Beijing). 2021 Jul;7(7):958-965. doi: 10.1016/j.eng.2021.04.015. Epub 2021 May 18. Engineering (Beijing). 2021. PMID: 34026297 Free PMC article.
-
First international external quality assessment of molecular diagnostics for Mers-CoV.J Clin Virol. 2015 Aug;69:81-5. doi: 10.1016/j.jcv.2015.05.022. Epub 2015 May 30. J Clin Virol. 2015. PMID: 26209385 Free PMC article.
-
Comparative Evaluation of Three Homogenization Methods for Isolating Middle East Respiratory Syndrome Coronavirus Nucleic Acids From Sputum Samples for Real-Time Reverse Transcription PCR.Ann Lab Med. 2016 Sep;36(5):457-62. doi: 10.3343/alm.2016.36.5.457. Ann Lab Med. 2016. PMID: 27374711 Free PMC article.
-
A case of imported COVID-19 diagnosed by PCR-positive lower respiratory specimen but with PCR-negative throat swabs.Infect Dis (Lond). 2020 Jun;52(6):423-426. doi: 10.1080/23744235.2020.1744711. Epub 2020 Apr 2. Infect Dis (Lond). 2020. PMID: 32238024 Free PMC article.
-
Influence of viral load in the outcome of hospitalized patients with influenza virus infection.Eur J Clin Microbiol Infect Dis. 2019 Apr;38(4):667-673. doi: 10.1007/s10096-019-03514-1. Epub 2019 Feb 28. Eur J Clin Microbiol Infect Dis. 2019. PMID: 30820840 Free PMC article.
References
-
- Hung IFN, Lau SKP, Woo PCY, et al. Viral loads in clinical specimens and SARS manifestations. Hong Kong Med J. 2009;15:S20–2. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

