RBPmap: a web server for mapping binding sites of RNA-binding proteins
- PMID: 24829458
- PMCID: PMC4086114
- DOI: 10.1093/nar/gku406
RBPmap: a web server for mapping binding sites of RNA-binding proteins
Abstract
Regulation of gene expression is executed in many cases by RNA-binding proteins (RBPs) that bind to mRNAs as well as to non-coding RNAs. RBPs recognize their RNA target via specific binding sites on the RNA. Predicting the binding sites of RBPs is known to be a major challenge. We present a new webserver, RBPmap, freely accessible through the website http://rbpmap.technion.ac.il/ for accurate prediction and mapping of RBP binding sites. RBPmap has been developed specifically for mapping RBPs in human, mouse and Drosophila melanogaster genomes, though it supports other organisms too. RBPmap enables the users to select motifs from a large database of experimentally defined motifs. In addition, users can provide any motif of interest, given as either a consensus or a PSSM. The algorithm for mapping the motifs is based on a Weighted-Rank approach, which considers the clustering propensity of the binding sites and the overall tendency of regulatory regions to be conserved. In addition, RBPmap incorporates a position-specific background model, designed uniquely for different genomic regions, such as splice sites, 5' and 3' UTRs, non-coding RNA and intergenic regions. RBPmap was tested on high-throughput RNA-binding experiments and was proved to be highly accurate.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
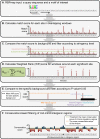
Figures

Similar articles
-
RBPmap: A Tool for Mapping and Predicting the Binding Sites of RNA-Binding Proteins Considering the Motif Environment.Methods Mol Biol. 2022;2404:53-65. doi: 10.1007/978-1-0716-1851-6_3. Methods Mol Biol. 2022. PMID: 34694603
-
SMARTIV: combined sequence and structure de-novo motif discovery for in-vivo RNA binding data.Nucleic Acids Res. 2018 Jul 2;46(W1):W221-W228. doi: 10.1093/nar/gky453. Nucleic Acids Res. 2018. PMID: 29800452 Free PMC article.
-
BRIO: a web server for RNA sequence and structure motif scan.Nucleic Acids Res. 2021 Jul 2;49(W1):W67-W71. doi: 10.1093/nar/gkab400. Nucleic Acids Res. 2021. PMID: 34038531 Free PMC article.
-
Finding the target sites of RNA-binding proteins.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):111-30. doi: 10.1002/wrna.1201. Epub 2013 Nov 11. Wiley Interdiscip Rev RNA. 2014. PMID: 24217996 Free PMC article. Review.
-
RNA-protein interactions: an overview.Methods Mol Biol. 2014;1097:491-521. doi: 10.1007/978-1-62703-709-9_23. Methods Mol Biol. 2014. PMID: 24639174 Review.
Cited by
-
A massively parallel reporter assay reveals focused and broadly encoded RNA localization signals in neurons.Nucleic Acids Res. 2022 Oct 14;50(18):10643-10664. doi: 10.1093/nar/gkac806. Nucleic Acids Res. 2022. PMID: 36156153 Free PMC article.
-
The RNA-binding protein PTBP1 promotes ATPase-dependent dissociation of the RNA helicase UPF1 to protect transcripts from nonsense-mediated mRNA decay.J Biol Chem. 2020 Aug 14;295(33):11613-11625. doi: 10.1074/jbc.RA120.013824. Epub 2020 Jun 22. J Biol Chem. 2020. PMID: 32571872 Free PMC article.
-
Alternative splicing of U2AF1 reveals a shared repression mechanism for duplicated exons.Nucleic Acids Res. 2017 Jan 9;45(1):417-434. doi: 10.1093/nar/gkw733. Epub 2016 Aug 26. Nucleic Acids Res. 2017. PMID: 27566151 Free PMC article.
-
Pervasive isoform-specific translational regulation via alternative transcription start sites in mammals.Mol Syst Biol. 2016 Jul 18;12(7):875. doi: 10.15252/msb.20166941. Mol Syst Biol. 2016. PMID: 27430939 Free PMC article.
-
Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer.J Adv Res. 2021 Oct 5;37:91-106. doi: 10.1016/j.jare.2021.10.001. eCollection 2022 Mar. J Adv Res. 2021. PMID: 35499052 Free PMC article.
References
-
- Ray D., Kazan H., Chan E.T., Peña Castillo L., Chaudhry S., Talukder S., Blencowe B.J., Morris Q., Hughes T.R. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009;27:667–670. - PubMed
-
- Keene J.D., Komisarow J.M., Friedersdorf M.B. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006;1:302–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

