Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon
- PMID: 24820376
- PMCID: PMC4126194
- DOI: 10.1038/nmeth.2889
Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon
Abstract
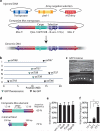
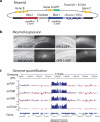
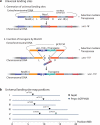
We have generated a recombinant Mos1 transposon that can insert up to 45-kb transgenes into the Caenorhabditis elegans genome. The minimal Mos1 transposon (miniMos) is 550 bp long and inserts DNA into the genome at high frequency (~60% of injected animals). Genetic and antibiotic markers can be used for selection, and the transposon is active in C. elegans isolates and Caenorhabditis briggsae. We used the miniMos transposon to generate six universal Mos1-mediated single-copy insertion (mosSCI) landing sites that allow targeted transgene insertion with a single targeting vector into permissive expression sites on all autosomes. We also generated two collections of strains: a set of bright fluorescent insertions that are useful as dominant, genetic balancers and a set of lacO insertions to track genome position.
Figures
Similar articles
-
Transposon-Assisted Genetic Engineering with Mos1-Mediated Single-Copy Insertion (MosSCI).Methods Mol Biol. 2015;1327:49-58. doi: 10.1007/978-1-4939-2842-2_5. Methods Mol Biol. 2015. PMID: 26423967
-
Engineering the Caenorhabditis elegans genome by Mos1-induced transgene-instructed gene conversion.Methods Mol Biol. 2012;859:189-201. doi: 10.1007/978-1-61779-603-6_11. Methods Mol Biol. 2012. PMID: 22367873
-
Targeted and Random Transposon-Assisted Single-Copy Transgene Insertion in C. elegans.Methods Mol Biol. 2022;2468:239-256. doi: 10.1007/978-1-0716-2181-3_12. Methods Mol Biol. 2022. PMID: 35320568
-
Insertional mutagenesis in C. elegans using the Drosophila transposon Mos1: a method for the rapid identification of mutated genes.Methods Mol Biol. 2006;351:59-73. doi: 10.1385/1-59745-151-7:59. Methods Mol Biol. 2006. PMID: 16988426 Review.
-
The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans.Genetica. 2009 Sep;137(1):39-46. doi: 10.1007/s10709-009-9361-3. Epub 2009 Apr 3. Genetica. 2009. PMID: 19343510 Review.
Cited by
-
The asymmetry of female meiosis reduces the frequency of inheritance of unpaired chromosomes.Elife. 2015 Apr 7;4:e06056. doi: 10.7554/eLife.06056. Elife. 2015. PMID: 25848744 Free PMC article.
-
Acute and inherited piRNA-mediated silencing in a rde-3 ribonucleotidyltransferase mutant.MicroPubl Biol. 2022 Sep 14;2022:10.17912/micropub.biology.000638. doi: 10.17912/micropub.biology.000638. eCollection 2022. MicroPubl Biol. 2022. PMID: 36188099 Free PMC article.
-
Targeted DamID in C. elegans reveals a direct role for LIN-22 and NHR-25 in antagonizing the epidermal stem cell fate.Sci Adv. 2022 Feb 4;8(5):eabk3141. doi: 10.1126/sciadv.abk3141. Epub 2022 Feb 4. Sci Adv. 2022. PMID: 35119932 Free PMC article.
-
The G2-to-M Transition Is Ensured by a Dual Mechanism that Protects Cyclin B from Degradation by Cdc20-Activated APC/C.Dev Cell. 2019 Nov 4;51(3):313-325.e10. doi: 10.1016/j.devcel.2019.09.005. Epub 2019 Oct 3. Dev Cell. 2019. PMID: 31588029 Free PMC article.
-
Influences of HLH-2 stability on anchor cell fate specification during Caenorhabditis elegans gonadogenesis.G3 (Bethesda). 2022 Apr 4;12(4):jkac028. doi: 10.1093/g3journal/jkac028. G3 (Bethesda). 2022. PMID: 35134193 Free PMC article.
References
-
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Dev. Camb. Engl. 1993;118:401–415. - PubMed
-
- Venken KJT, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. - PubMed
-
- Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

