The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans
- PMID: 24813612
- PMCID: PMC4454526
- DOI: 10.1016/j.cell.2014.02.055
The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans
Abstract
The increased longevity of the C. elegans electron transport chain mutants isp-1 and nuo-6 is mediated by mitochondrial ROS (mtROS) signaling. Here we show that the mtROS signal is relayed by the conserved, mitochondria-associated, intrinsic apoptosis signaling pathway (CED-9/Bcl2, CED-4/Apaf1, and CED-3/Casp9) triggered by CED-13, an alternative BH3-only protein. Activation of the pathway by an elevation of mtROS does not affect apoptosis but protects from the consequences of mitochondrial dysfunction by triggering a unique pattern of gene expression that modulates stress sensitivity and promotes survival. In vertebrates, mtROS induce apoptosis through the intrinsic pathway to protect from severely damaged cells. Our observations in nematodes demonstrate that sensing of mtROS by the apoptotic pathway can, independently of apoptosis, elicit protective mechanisms that keep the organism alive under stressful conditions. This results in extended longevity when mtROS generation is inappropriately elevated. These findings clarify the relationships between mitochondria, ROS, apoptosis, and aging.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
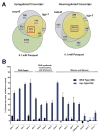
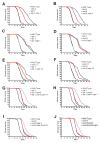
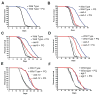
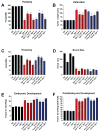
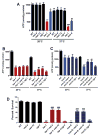
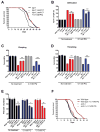
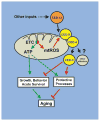
Comment in
-
Aging: the blurry line between life and death.Curr Biol. 2014 Jul 7;24(13):R610-3. doi: 10.1016/j.cub.2014.05.057. Curr Biol. 2014. PMID: 25004366
Similar articles
-
A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans.PLoS Biol. 2010 Dec 7;8(12):e1000556. doi: 10.1371/journal.pbio.1000556. PLoS Biol. 2010. PMID: 21151885 Free PMC article.
-
Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans.Aging Cell. 2010 Jun;9(3):433-47. doi: 10.1111/j.1474-9726.2010.00571.x. Epub 2010 Mar 19. Aging Cell. 2010. PMID: 20346072
-
Uncoupling of oxidative stress resistance and lifespan in long-lived isp-1 mitochondrial mutants in Caenorhabditis elegans.Free Radic Biol Med. 2017 Jul;108:362-373. doi: 10.1016/j.freeradbiomed.2017.04.004. Epub 2017 Apr 7. Free Radic Biol Med. 2017. PMID: 28392283 Free PMC article.
-
Mitochondrial dysfunction and longevity in animals: Untangling the knot.Science. 2015 Dec 4;350(6265):1204-7. doi: 10.1126/science.aac4357. Science. 2015. PMID: 26785479 Review.
-
Effects of the mitochondrial respiratory chain on longevity in C. elegans.Exp Gerontol. 2014 Aug;56:245-55. doi: 10.1016/j.exger.2014.03.028. Epub 2014 Apr 5. Exp Gerontol. 2014. PMID: 24709342 Review.
Cited by
-
Human exceptional longevity: transcriptome from centenarians is distinct from septuagenarians and reveals a role of Bcl-xL in successful aging.Aging (Albany NY). 2016 Oct 28;8(12):3185-3208. doi: 10.18632/aging.101078. Aging (Albany NY). 2016. PMID: 27794564 Free PMC article.
-
Axonal Mitochondria Modulate Neuropeptide Secretion Through the Hypoxic Stress Response in Caenorhabditis elegans.Genetics. 2018 Sep;210(1):275-285. doi: 10.1534/genetics.118.301014. Epub 2018 Jul 26. Genetics. 2018. PMID: 30049781 Free PMC article.
-
The Neuroprotector Benzothiazepine CGP37157 Extends Lifespan in C. elegans Worms.Front Aging Neurosci. 2019 Jan 17;10:440. doi: 10.3389/fnagi.2018.00440. eCollection 2018. Front Aging Neurosci. 2019. PMID: 30705628 Free PMC article.
-
Caloric Restriction Induces MicroRNAs to Improve Mitochondrial Proteostasis.iScience. 2019 Jul 26;17:155-166. doi: 10.1016/j.isci.2019.06.028. Epub 2019 Jun 21. iScience. 2019. PMID: 31279933 Free PMC article.
-
A bacterial pathogen induces developmental slowing by high reactive oxygen species and mitochondrial dysfunction in Caenorhabditis elegans.Cell Rep. 2023 Oct 31;42(10):113189. doi: 10.1016/j.celrep.2023.113189. Epub 2023 Oct 5. Cell Rep. 2023. PMID: 37801396 Free PMC article.
References
-
- Baruah HC, Hall Mathew, Yuan Jie, Gordon Sarah, Johnson Erik, Shtessel Ludmila L, Yee Callista, Hekimi Siegfried, Brent Derry W, Lee Siu Sylvia. CEP-1, the Caenorhabditis elegans p53 homolog, mediates opposing longevity outcomes in mitochondrial electron transport chain mutants. PLoS genetics 2014 - PMC - PubMed
-
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. - PubMed
-
- Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes & development. 2008;22:3236–3241. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

