Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates
- PMID: 24811749
- PMCID: PMC4193922
- DOI: 10.1002/embj.201386906
Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates
Abstract
Degradation rates of most proteins in eukaryotic cells are determined by their rates of ubiquitination. However, possible regulation of the proteasome's capacity to degrade ubiquitinated proteins has received little attention, although proteasome inhibitors are widely used in research and cancer treatment. We show here that mammalian 26S proteasomes have five associated ubiquitin ligases and that multiple proteasome subunits are ubiquitinated in cells, especially the ubiquitin receptor subunit, Rpn13. When proteolysis is even partially inhibited in cells or purified 26S proteasomes with various inhibitors, Rpn13 becomes extensively and selectively poly-ubiquitinated by the proteasome-associated ubiquitin ligase, Ube3c/Hul5. This modification also occurs in cells during heat-shock or arsenite treatment, when poly-ubiquitinated proteins accumulate. Rpn13 ubiquitination strongly decreases the proteasome's ability to bind and degrade ubiquitin-conjugated proteins, but not its activity against peptide substrates. This autoinhibitory mechanism presumably evolved to prevent binding of ubiquitin conjugates to defective or stalled proteasomes, but this modification may also be useful as a biomarker indicating the presence of proteotoxic stress and reduced proteasomal capacity in cells or patients.
Keywords: 26S proteasomes; Ube3c/Hul5; proteasome inhibitors; proteasome regulation; ubiquitination.
© 2014 The Authors.
Figures

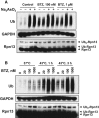
HEK293F parental cells and cells stably overexpressing Dss1-FLAG were treated with different concentrations of BTZ for various times. Levels of proteasomes and ligases in the crude extracts were analyzed by Western blot.
Proteasomes were isolated from cells generated in (A) using anti-FLAG affinity resin. Equal amounts of proteasomes were analyzed by Western blot. Huwe1 was not detectable under these conditions, most likely due to the small amounts of cells used in this experiment compared to those used in our initial purifications (e.g. Supplementary Fig S2C).
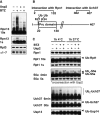
26S proteasomes were isolated from Dss1-FLAG-expressing HEK293F cells that were treated with or without 1 μM BTZ for 4 h, and ubiquitination of different subunits analyzed by Western blot (WB). To confirm that these subunits were modified by ubiquitination, these particles were incubated with the deubiquitinating enzyme Usp2 (1 μM) for 1 h at 4°C.
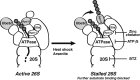
Schematic view of Rpn13. K34 is located in, and K21 adjacent to the Pru domain responsible for binding of Ub conjugates.
Proteasomes isolated from normal and BTZ-treated cells were treated with 1 μM Usp2 for 1 h at either 4°C or 37°C. Samples were analyzed by Western blot.
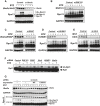
Rpn13 ubiquitination was stimulated by inhibiting 20S function with BTZ, 19S ATPases with ATPγS, or Rpn11 with a Zn2+-chelating agent. Isolated proteasomes were incubated at 37°C for the times indicated in the presence of ATP, Ub, E1, and Ubch5a as the E2. Rpn13 ubiquitination was then analyzed by WB. Left panel, ubiquitination carried out with or without BTZ. Central panel, ATP was substituted with ATPγS where indicated. Right panel, ubiquitination carried out with or without a zinc-chelating agent.
Only Rpn13 is extensively modified by poly-ubiquitination. Isolated 26S proteasomes were incubated in the presence of ATP, Ub, and E1 with or without the E2 for 2 h at 37°C and analyzed by Western blot. BTZ and Ub aldehyde were included in all reactions.
Use of single-lysine Ub mutants or Ub mutants lacking specific lysines indicates that Rpn13 was poly-ubiquitinated by predominantly K29 and K48-linked Ub chains. Proteasomes were ubiquitinated as in (A) and (B) with or without wild-type or mutant Ub as indicated, and Rpn13 ubiquitination was analyzed by Western blot. Top panel, Ub point mutants. Bottom panel, single-lysine Ub mutants.
No deubiquitination of immobilized 26S proteasomes (FLAG) is observed after removing the ubiquitination mixture (S/N) and replacing it with buffer, followed by incubation at 37°C. Usp2 treatment readily deubiquitinates Rpn13 under these conditions.
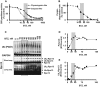
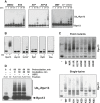
HEK293F cells were treated with 0.5 μM Na3AsO3 together with indicated concentrations of BTZ for 3 h. Na3AsO3 dramatically caused the accumulation of poly-ubiquitinated proteins and induced Rpn13 ubiquitination as well.
HEK293F cells were treated with increasing concentrations of BTZ for 3 h at 37°C or under 43°C (heat shock) for 1 or 3 h. Heat shock and arsenite also caused the accumulation of poly-ubiquitinated proteins as well as Rpn13 ubiquitination.
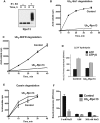
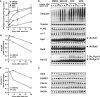
Isolated proteasomes were preincubated for 0–2 h at 37°C in the presence of ATP and Ub, with or without E1 and Ubch5a (E2) as indicated and analyzed by Western blot. (Ub aldehyde was included in all reactions to block deubiquitination, but its presence had no significant effect.)
Proteasomes preincubated for 2 h as in (A) were diluted 20-fold, and the degradation of 32P-labeled Ubn-Sic1 was assayed by measuring the production of TCA-soluble radioactive peptides.
After preincubation, degradation of 32P-labeled Ub5-DHFR was assayed as in (B).
To evaluate the effects of Rpn13 modification on hydrolysis of short peptides, proteasomes were preincubated for 2 h as in (A) and diluted 20-fold, and the rate of suc-LLVY-amc hydrolysis was measured in the presence of ATP or ATPγS, which stimulates gate opening and peptide substrate entry (Smith et al, 2007).
To follow the degradation of a non-ubiquitinated substrate, 26S proteasomes were preincubated as in (A), and the breakdown of 32P-labeled casein to TCA-soluble peptides was assayed as in (B).
To measure the binding of ubiquitinated proteins, proteasomes were preincubated for 2 h as in (A) and bound to immobilized Ub5-DHFR at 4°C. The resin was washed in a low-salt buffer (0 mM NaCl), high-salt buffer (300 mM NaCl) or in the presence of UIM, and bound proteasome activity was assayed by measuring suc-LLVY-amc hydrolysis (Peth et al, 2010).
Similar articles
-
Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c.Proc Natl Acad Sci U S A. 2017 Apr 25;114(17):E3404-E3413. doi: 10.1073/pnas.1701734114. Epub 2017 Apr 10. Proc Natl Acad Sci U S A. 2017. PMID: 28396413 Free PMC article.
-
Autoregulation of the 26S proteasome by in situ ubiquitination.Mol Biol Cell. 2014 Jun 15;25(12):1824-35. doi: 10.1091/mbc.E13-10-0585. Epub 2014 Apr 17. Mol Biol Cell. 2014. PMID: 24743594 Free PMC article.
-
26S proteasomes become stably activated upon heat shock when ubiquitination and protein degradation increase.Proc Natl Acad Sci U S A. 2022 Jun 21;119(25):e2122482119. doi: 10.1073/pnas.2122482119. Epub 2022 Jun 15. Proc Natl Acad Sci U S A. 2022. PMID: 35704754 Free PMC article.
-
[Ubiquitin-independent protein degradation in proteasomes].Biomed Khim. 2018 Mar;64(2):134-148. doi: 10.18097/PBMC20186402134. Biomed Khim. 2018. PMID: 29723144 Review. Russian.
-
Mechanisms That Activate 26S Proteasomes and Enhance Protein Degradation.Biomolecules. 2021 May 22;11(6):779. doi: 10.3390/biom11060779. Biomolecules. 2021. PMID: 34067263 Free PMC article. Review.
Cited by
-
The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death.Cell Res. 2016 Aug;26(8):869-85. doi: 10.1038/cr.2016.86. Epub 2016 Jul 22. Cell Res. 2016. PMID: 27444871 Free PMC article. Review.
-
The Ubiquitination System within Bacterial Host-Pathogen Interactions.Microorganisms. 2021 Mar 19;9(3):638. doi: 10.3390/microorganisms9030638. Microorganisms. 2021. PMID: 33808578 Free PMC article. Review.
-
Pharmacologically increasing cGMP improves proteostasis and reduces neuropathy in mouse models of CMT1.Cell Mol Life Sci. 2024 Oct 14;81(1):434. doi: 10.1007/s00018-024-05463-1. Cell Mol Life Sci. 2024. PMID: 39400753 Free PMC article.
-
Ubiquitination-mediated molecular pathway alterations in human lung squamous cell carcinomas identified by quantitative ubiquitinomics.Front Endocrinol (Lausanne). 2022 Sep 15;13:970843. doi: 10.3389/fendo.2022.970843. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36187110 Free PMC article.
-
The Multifaceted Roles of USP15 in Signal Transduction.Int J Mol Sci. 2021 Apr 29;22(9):4728. doi: 10.3390/ijms22094728. Int J Mol Sci. 2021. PMID: 33946990 Free PMC article. Review.
References
-
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

