Determinants of substrate and cation transport in the human Na+/dicarboxylate cotransporter NaDC3
- PMID: 24808185
- PMCID: PMC4059142
- DOI: 10.1074/jbc.M114.554790
Determinants of substrate and cation transport in the human Na+/dicarboxylate cotransporter NaDC3
Abstract
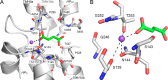
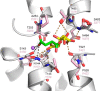
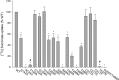
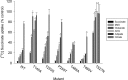
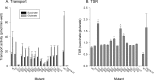
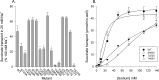
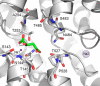
Metabolic intermediates, such as succinate and citrate, regulate important processes ranging from energy metabolism to fatty acid synthesis. Cytosolic concentrations of these metabolites are controlled, in part, by members of the SLC13 gene family. The molecular mechanism underlying Na(+)-coupled di- and tricarboxylate transport by this family is understood poorly. The human Na(+)/dicarboxylate cotransporter NaDC3 (SLC13A3) is found in various tissues, including the kidney, liver, and brain. In addition to citric acid cycle intermediates such as α-ketoglutarate and succinate, NaDC3 transports other compounds into cells, including N-acetyl aspartate, mercaptosuccinate, and glutathione, in keeping with its dual roles in cell nutrition and detoxification. In this study, we construct a homology structural model of NaDC3 on the basis of the structure of the Vibrio cholerae homolog vcINDY. Our computations are followed by experimental testing of the predicted NaDC3 structure and mode of interaction with various substrates. The results of this study show that the substrate and cation binding domains of NaDC3 are composed of residues in the opposing hairpin loops and unwound portions of adjacent helices. Furthermore, these results provide a possible explanation for the differential substrate specificity among dicarboxylate transporters that underpin their diverse biological roles in metabolism and detoxification. The structural model of NaDC3 provides a framework for understanding substrate selectivity and the Na(+)-coupled anion transport mechanism by the human SLC13 family and other key solute carrier transporters.
Keywords: Citrate; Homology Modeling; Membrane; Molecular Docking; SLC13 Family; Sodium Transport; Succinate; Tricarboxylic Acid Cycle (TCA Cycle) (Krebs Cycle).
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

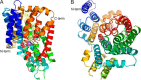
Similar articles
-
Sodium-coupled dicarboxylate and citrate transporters from the SLC13 family.Pflugers Arch. 2014 Jan;466(1):119-30. doi: 10.1007/s00424-013-1369-y. Epub 2013 Oct 10. Pflugers Arch. 2014. PMID: 24114175 Review.
-
Mapping Functionally Important Residues in the Na+/Dicarboxylate Cotransporter, NaDC1.Biochemistry. 2017 Aug 22;56(33):4432-4441. doi: 10.1021/acs.biochem.7b00503. Epub 2017 Aug 10. Biochemistry. 2017. PMID: 28731330
-
Functional and molecular identification of sodium-coupled dicarboxylate transporters in rat primary cultured cerebrocortical astrocytes and neurons.J Neurochem. 2006 Apr;97(1):162-73. doi: 10.1111/j.1471-4159.2006.03720.x. Epub 2006 Mar 8. J Neurochem. 2006. PMID: 16524379
-
Threonine-509 is a determinant of apparent affinity for both substrate and cations in the human Na+/dicarboxylate cotransporter.Biochemistry. 2008 Jan 22;47(3):1087-93. doi: 10.1021/bi701417h. Epub 2007 Dec 28. Biochemistry. 2008. PMID: 18161988 Free PMC article.
-
Role of sodium dependent SLC13 transporter inhibitors in various metabolic disorders.Mol Cell Biochem. 2023 Aug;478(8):1669-1687. doi: 10.1007/s11010-022-04618-7. Epub 2022 Dec 10. Mol Cell Biochem. 2023. PMID: 36495372 Review.
Cited by
-
Mutations in the Na(+)/citrate cotransporter NaCT (SLC13A5) in pediatric patients with epilepsy and developmental delay.Mol Med. 2016 May 26;22:310-21. doi: 10.2119/molmed.2016.00077. Mol Med. 2016. PMID: 27261973 Free PMC article.
-
Analysis of naturally occurring mutations in the human uptake transporter NaCT important for bone and brain development and energy metabolism.Sci Rep. 2018 Jul 27;8(1):11330. doi: 10.1038/s41598-018-29547-8. Sci Rep. 2018. PMID: 30054523 Free PMC article.
-
Focally administered succinate improves cerebral metabolism in traumatic brain injury patients with mitochondrial dysfunction.J Cereb Blood Flow Metab. 2022 Jan;42(1):39-55. doi: 10.1177/0271678X211042112. Epub 2021 Sep 8. J Cereb Blood Flow Metab. 2022. PMID: 34494481 Free PMC article. Clinical Trial.
-
Focally perfused succinate potentiates brain metabolism in head injury patients.J Cereb Blood Flow Metab. 2017 Jul;37(7):2626-2638. doi: 10.1177/0271678X16672665. Epub 2016 Jan 1. J Cereb Blood Flow Metab. 2017. PMID: 27798266 Free PMC article.
-
Structure-Based Identification of Inhibitors for the SLC13 Family of Na(+)/Dicarboxylate Cotransporters.Biochemistry. 2015 Aug 11;54(31):4900-8. doi: 10.1021/acs.biochem.5b00388. Epub 2015 Jul 30. Biochemistry. 2015. PMID: 26176240 Free PMC article.
References
-
- Gullans S. R., Kone B. C., Avison M. J., Giebisch G. (1988) Succinate alters respiration, membrane potential, and intracellular K+ in proximal tubule. Am. J. Physiol. 255, F1170-F1177 - PubMed
-
- Ruderman N. B., Saha A. K., Vavvas D., Witters L. A. (1999) Malonyl-CoA, fuel sensing, and insulin resistance. Am. J. Physiol. 276, E1–E18 - PubMed
-
- Stoppa G. R., Cesquini M., Roman E. A., Prada P. O., Torsoni A. S., Romanatto T., Saad M. J., Velloso L. A., Torsoni M. A. (2008) Intracerebroventricular injection of citrate inhibits hypothalamic AMPK and modulates feeding behavior and peripheral insulin signaling. J. Endocrinol. 198, 157–168 - PubMed
-
- Pajor A. M. (2014) Sodium-coupled dicarboxylate and citrate transporters from the SLC13 family. Pflugers Arch. 466, 119–130 - PubMed
-
- Rogina B., Reenan R. A., Nilsen S. P., Helfand S. L. (2000) Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290, 2137–2140 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

