C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD
- PMID: 24806409
- PMCID: PMC4161616
- DOI: 10.1007/s00401-014-1286-y
C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD
Abstract
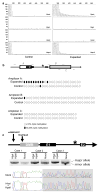
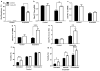
Hexanucleotide repeat expansions of C9orf72 are the most common genetic cause of amyotrophic lateral sclerosis and frontotemporal degeneration. The mutation is associated with reduced C9orf72 expression and the accumulation of potentially toxic RNA and protein aggregates. CpG methylation is known to protect the genome against unstable DNA elements and to stably silence inappropriate gene expression. Using bisulfite cloning and restriction enzyme-based methylation assays on DNA from human brain and peripheral blood, we observed CpG hypermethylation involving the C9orf72 promoter in cis to the repeat expansion mutation in approximately one-third of C9orf72 repeat expansion mutation carriers. Promoter hypermethylation of mutant C9orf72 was associated with transcriptional silencing of C9orf72 in patient-derived lymphoblast cell lines, resulting in reduced accumulation of intronic C9orf72 RNA and reduced numbers of RNA foci. Furthermore, demethylation of mutant C9orf72 with 5-aza-deoxycytidine resulted in increased vulnerability of mutant cells to oxidative and autophagic stress. Promoter hypermethylation of repeat expansion carriers was also associated with reduced accumulation of RNA foci and dipeptide repeat protein aggregates in human brains. These results indicate that C9orf72 promoter hypermethylation prevents downstream molecular aberrations associated with the hexanucleotide repeat expansion, suggesting that epigenetic silencing of the mutant C9orf72 allele may represent a protective counter-regulatory response to hexanucleotide repeat expansion.
Figures
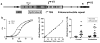
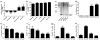
Similar articles
-
Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier.Acta Neuropathol. 2015 Jan;129(1):39-52. doi: 10.1007/s00401-014-1365-0. Epub 2014 Nov 12. Acta Neuropathol. 2015. PMID: 25388784 Free PMC article.
-
Elevated methylation levels, reduced expression levels, and frequent contractions in a clinical cohort of C9orf72 expansion carriers.Mol Neurodegener. 2020 Jan 30;15(1):7. doi: 10.1186/s13024-020-0359-8. Mol Neurodegener. 2020. PMID: 32000838 Free PMC article.
-
Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice.Neuron. 2015 Dec 2;88(5):902-909. doi: 10.1016/j.neuron.2015.11.018. Neuron. 2015. PMID: 26637797 Free PMC article.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
-
Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene.Neurosci Lett. 2017 Jan 1;636:16-26. doi: 10.1016/j.neulet.2016.09.007. Epub 2016 Sep 13. Neurosci Lett. 2017. PMID: 27619540 Free PMC article. Review.
Cited by
-
Semi-automated quantification of C9orf72 expansion size reveals inverse correlation between hexanucleotide repeat number and disease duration in frontotemporal degeneration.Acta Neuropathol. 2015 Sep;130(3):363-72. doi: 10.1007/s00401-015-1445-9. Epub 2015 May 29. Acta Neuropathol. 2015. PMID: 26022924 Free PMC article.
-
Biomarkers in Motor Neuron Disease: A State of the Art Review.Front Neurol. 2019 Apr 3;10:291. doi: 10.3389/fneur.2019.00291. eCollection 2019. Front Neurol. 2019. PMID: 31001186 Free PMC article. Review.
-
Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair.Hum Mol Genet. 2020 Aug 3;29(13):2200-2217. doi: 10.1093/hmg/ddaa106. Hum Mol Genet. 2020. PMID: 32504093 Free PMC article.
-
Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72.Nat Neurosci. 2020 May;23(5):615-624. doi: 10.1038/s41593-020-0619-5. Epub 2020 Apr 13. Nat Neurosci. 2020. PMID: 32284607 Free PMC article.
-
Neurodegeneration in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9orf72 is linked to TDP-43 pathology and not associated with aggregated forms of dipeptide repeat proteins.Neuropathol Appl Neurobiol. 2016 Apr;42(3):242-54. doi: 10.1111/nan.12292. Epub 2015 Dec 7. Neuropathol Appl Neurobiol. 2016. PMID: 26538301 Free PMC article.
References
-
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, Seeley WW, Boxer AL, Petrucelli L, Miller BL, Gao FB. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126(3):385–399. doi: 10.1007/s00401-013-1149-y. - DOI - PMC - PubMed
-
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. - DOI - PMC - PubMed
-
- Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, Boylan K, Patel TC, Dickson DW, Petrucelli L. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethyla-tion, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126(6):895–905. doi: 10.1007/s00401-013-1199-1. - DOI - PMC - PubMed
-
- Benussi L, Rossi G, Glionna M, Tonoli E, Piccoli E, Fostinelli S, Paterlini A, Flocco R, Albani D, Pantieri R, Cereda C, Forloni G, Tagliavini F, Binetti G, Ghidoni R. C9ORF72 hexanucleotide repeat number in frontotemporal lobar degeneration: a genotype–phenotype correlation study. J Alzheimers Dis. 2013 doi: 10.3233/JAD-131028. - DOI - PubMed
-
- Boeve BF, Boylan KB, Graff-Radford NR, DeJesus-Hernandez M, Knopman DS, Pedraza O, Vemuri P, Jones D, Lowe V, Murray ME, Dickson DW, Josephs KA, Rush BK, Machulda MM, Fields JA, Ferman TJ, Baker M, Rutherford NJ, Adamson J, Wszolek ZK, Adeli A, Savica R, Boot B, Kuntz KM, Gavrilova R, Reeves A, Whitwell J, Kantarci K, Jack CR, Jr, Parisi JE, Lucas JA, Petersen RC, Rademakers R. Characterization of fron-totemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain. 2012;135(Pt 3):765–783. doi: 10.1093/brain/aws004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG032953/AG/NIA NIH HHS/United States
- T32AG00255/AG/NIA NIH HHS/United States
- P01 AG017586/AG/NIA NIH HHS/United States
- T32 AG000255/AG/NIA NIH HHS/United States
- K08 AG039510/AG/NIA NIH HHS/United States
- P01AG017586/AG/NIA NIH HHS/United States
- P30AG10125/AG/NIA NIH HHS/United States
- P01AG032953/AG/NIA NIH HHS/United States
- ND14442/ND/ONDIEH CDC HHS/United States
- K08AG039510/AG/NIA NIH HHS/United States
- P30 AG010124/AG/NIA NIH HHS/United States
- ND10966/ND/ONDIEH CDC HHS/United States
- ND16183/ND/ONDIEH CDC HHS/United States
- K23 NS088341/NS/NINDS NIH HHS/United States
- ND11836/ND/ONDIEH CDC HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

