Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis
- PMID: 24788150
- PMCID: PMC4006877
- DOI: 10.1371/journal.ppat.1004077
Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis
Abstract
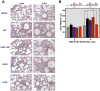
Deletion of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) envelope (E) gene attenuates the virus. E gene encodes a small multifunctional protein that possesses ion channel (IC) activity, an important function in virus-host interaction. To test the contribution of E protein IC activity in virus pathogenesis, two recombinant mouse-adapted SARS-CoVs, each containing one single amino acid mutation that suppressed ion conductivity, were engineered. After serial infections, mutant viruses, in general, incorporated compensatory mutations within E gene that rendered active ion channels. Furthermore, IC activity conferred better fitness in competition assays, suggesting that ion conductivity represents an advantage for the virus. Interestingly, mice infected with viruses displaying E protein IC activity, either with the wild-type E protein sequence or with the revertants that restored ion transport, rapidly lost weight and died. In contrast, mice infected with mutants lacking IC activity, which did not incorporate mutations within E gene during the experiment, recovered from disease and most survived. Knocking down E protein IC activity did not significantly affect virus growth in infected mice but decreased edema accumulation, the major determinant of acute respiratory distress syndrome (ARDS) leading to death. Reduced edema correlated with lung epithelia integrity and proper localization of Na+/K+ ATPase, which participates in edema resolution. Levels of inflammasome-activated IL-1β were reduced in the lung airways of the animals infected with viruses lacking E protein IC activity, indicating that E protein IC function is required for inflammasome activation. Reduction of IL-1β was accompanied by diminished amounts of TNF and IL-6 in the absence of E protein ion conductivity. All these key cytokines promote the progression of lung damage and ARDS pathology. In conclusion, E protein IC activity represents a new determinant for SARS-CoV virulence.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis.mBio. 2018 May 22;9(3):e02325-17. doi: 10.1128/mBio.02325-17. mBio. 2018. PMID: 29789363 Free PMC article.
-
The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis.PLoS Pathog. 2014 Aug 14;10(8):e1004320. doi: 10.1371/journal.ppat.1004320. eCollection 2014 Aug. PLoS Pathog. 2014. PMID: 25122212 Free PMC article.
-
Coronavirus virulence genes with main focus on SARS-CoV envelope gene.Virus Res. 2014 Dec 19;194:124-37. doi: 10.1016/j.virusres.2014.07.024. Epub 2014 Aug 2. Virus Res. 2014. PMID: 25093995 Free PMC article. Review.
-
Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates.J Virol. 2015 Apr;89(7):3870-87. doi: 10.1128/JVI.03566-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609816 Free PMC article.
-
SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium.Virus Res. 2008 Apr;133(1):33-44. doi: 10.1016/j.virusres.2007.03.013. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451829 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Viroporin E Induces Ca2+ Release and Neuron Cell Death in Primary Cultures of Rat Hippocampal Cells Aged In Vitro.Int J Mol Sci. 2024 Jun 7;25(12):6304. doi: 10.3390/ijms25126304. Int J Mol Sci. 2024. PMID: 38928009 Free PMC article.
-
Novel Coronavirus-Induced NLRP3 Inflammasome Activation: A Potential Drug Target in the Treatment of COVID-19.Front Immunol. 2020 May 19;11:1021. doi: 10.3389/fimmu.2020.01021. eCollection 2020. Front Immunol. 2020. PMID: 32574259 Free PMC article. No abstract available.
-
Highlight of Immune Pathogenic Response and Hematopathologic Effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 Infection.Front Immunol. 2020 May 12;11:1022. doi: 10.3389/fimmu.2020.01022. eCollection 2020. Front Immunol. 2020. PMID: 32574260 Free PMC article. Review.
-
Characterization of the SARS-CoV-2 E Protein: Sequence, Structure, Viroporin, and Inhibitors.Protein Sci. 2021 Jun;30(6):1114-1130. doi: 10.1002/pro.4075. Epub 2021 Apr 13. Protein Sci. 2021. PMID: 33813796 Free PMC article. Review.
-
Challenges and prospects of COVID-19 vaccine development based on the progress made in SARS and MERS vaccine development.Transbound Emerg Dis. 2021 May;68(3):1111-1124. doi: 10.1111/tbed.13804. Epub 2020 Sep 23. Transbound Emerg Dis. 2021. PMID: 32815655 Free PMC article. Review.
References
-
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campganoli R, et al. (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300: 1394–1399. - PubMed
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, et al. (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348: 1967–1976. - PubMed
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367: 1814–1820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

