Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation
- PMID: 24787765
- PMCID: PMC4006889
- DOI: 10.1371/journal.ppat.1004058
Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation
Abstract
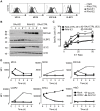
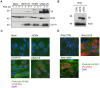
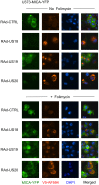
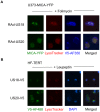
NKG2D plays a major role in controlling immune responses through the regulation of natural killer (NK) cells, αβ and γδ T-cell function. This activating receptor recognizes eight distinct ligands (the MHC Class I polypeptide-related sequences (MIC) A andB, and UL16-binding proteins (ULBP)1-6) induced by cellular stress to promote recognition cells perturbed by malignant transformation or microbial infection. Studies into human cytomegalovirus (HCMV) have aided both the identification and characterization of NKG2D ligands (NKG2DLs). HCMV immediate early (IE) gene up regulates NKGDLs, and we now describe the differential activation of ULBP2 and MICA/B by IE1 and IE2 respectively. Despite activation by IE functions, HCMV effectively suppressed cell surface expression of NKGDLs through both the early and late phases of infection. The immune evasion functions UL16, UL142, and microRNA(miR)-UL112 are known to target NKG2DLs. While infection with a UL16 deletion mutant caused the expected increase in MICB and ULBP2 cell surface expression, deletion of UL142 did not have a similar impact on its target, MICA. We therefore performed a systematic screen of the viral genome to search of addition functions that targeted MICA. US18 and US20 were identified as novel NK cell evasion functions capable of acting independently to promote MICA degradation by lysosomal degradation. The most dramatic effect on MICA expression was achieved when US18 and US20 acted in concert. US18 and US20 are the first members of the US12 gene family to have been assigned a function. The US12 family has 10 members encoded sequentially through US12-US21; a genetic arrangement, which is suggestive of an 'accordion' expansion of an ancestral gene in response to a selective pressure. This expansion must have be an ancient event as the whole family is conserved across simian cytomegaloviruses from old world monkeys. The evolutionary benefit bestowed by the combinatorial effect of US18 and US20 on MICA may have contributed to sustaining the US12 gene family.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein.J Immunol. 2003 Jul 15;171(2):902-8. doi: 10.4049/jimmunol.171.2.902. J Immunol. 2003. PMID: 12847260
-
Natural Killer Cell Evasion Is Essential for Infection by Rhesus Cytomegalovirus.PLoS Pathog. 2016 Aug 31;12(8):e1005868. doi: 10.1371/journal.ppat.1005868. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27580123 Free PMC article.
-
Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation.Elife. 2017 Feb 10;6:e22206. doi: 10.7554/eLife.22206. Elife. 2017. PMID: 28186488 Free PMC article.
-
The UL16-binding proteins, a novel family of MHC class I-related ligands for NKG2D, activate natural killer cell functions.Immunol Rev. 2001 Jun;181:185-92. doi: 10.1034/j.1600-065x.2001.1810115.x. Immunol Rev. 2001. PMID: 11513139 Review.
-
Release of Soluble Ligands for the Activating NKG2D Receptor: One More Immune Evasion Strategy Evolved by HIV-1 ?Curr Drug Targets. 2016;17(1):54-64. doi: 10.2174/1389450116666150630110329. Curr Drug Targets. 2016. PMID: 26122035 Review.
Cited by
-
A Portrait of the Human Organelle Proteome In Space and Time during Cytomegalovirus Infection.Cell Syst. 2016 Oct 26;3(4):361-373.e6. doi: 10.1016/j.cels.2016.08.012. Epub 2016 Sep 15. Cell Syst. 2016. PMID: 27641956 Free PMC article.
-
Dynamic Co-evolution of Host and Pathogen: HCMV Downregulates the Prevalent Allele MICA∗008 to Escape Elimination by NK Cells.Cell Rep. 2015 Feb 17;10(6):968-982. doi: 10.1016/j.celrep.2015.01.029. Epub 2015 Feb 12. Cell Rep. 2015. PMID: 25683719 Free PMC article.
-
High-throughput analysis of human cytomegalovirus genome diversity highlights the widespread occurrence of gene-disrupting mutations and pervasive recombination.J Virol. 2015 Aug 1;89(15):7673-7695. doi: 10.1128/JVI.00578-15. Epub 2015 May 13. J Virol. 2015. PMID: 25972543 Free PMC article.
-
High-Definition Analysis of Host Protein Stability during Human Cytomegalovirus Infection Reveals Antiviral Factors and Viral Evasion Mechanisms.Cell Host Microbe. 2018 Sep 12;24(3):447-460.e11. doi: 10.1016/j.chom.2018.07.011. Epub 2018 Aug 16. Cell Host Microbe. 2018. PMID: 30122656 Free PMC article.
-
The use of microRNA by human viruses: lessons from NK cells and HCMV infection.Semin Immunopathol. 2014 Nov;36(6):659-74. doi: 10.1007/s00281-014-0447-3. Epub 2014 Sep 19. Semin Immunopathol. 2014. PMID: 25234555 Review.
References
-
- Arvin AM, Fast P, Myers M, Plotkin S, Rabinovich R (2004) Vaccine development to prevent cytomegalovirus disease: report from the National Vaccine Advisory Committee. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 39: 233–239. - PubMed
-
- Hellstrand K, Martner A, Bergstrom T (2013) Valganciclovir in patients with glioblastoma. The New England journal of medicine 369: 2066. - PubMed
-
- Pierer M, Rothe K, Quandt D, Schulz A, Rossol M, et al. (2012) Association of anticytomegalovirus seropositivity with more severe joint destruction and more frequent joint surgery in rheumatoid arthritis. Arthritis and rheumatism 64: 1740–1749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 101835/WT_/Wellcome Trust/United Kingdom
- BB/F009836/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G1000236/MRC_/Medical Research Council/United Kingdom
- WT090323MA/WT_/Wellcome Trust/United Kingdom
- BBS/B/02525/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 100140/WT_/Wellcome Trust/United Kingdom
- MC_UU_12014/3/MRC_/Medical Research Council/United Kingdom
- MR/L018373/1/MRC_/Medical Research Council/United Kingdom
- G0700142/MRC_/Medical Research Council/United Kingdom
- MR/L008734/1/MRC_/Medical Research Council/United Kingdom
- G0901119/MRC_/Medical Research Council/United Kingdom
- 093966/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- G0801822/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

