DNA topology and transcription
- PMID: 24755522
- PMCID: PMC4133214
- DOI: 10.4161/nucl.28909
DNA topology and transcription
Abstract
Chromatin is a complex assembly that compacts DNA inside the nucleus while providing the necessary level of accessibility to regulatory factors conscripted by cellular signaling systems. In this superstructure, DNA is the subject of mechanical forces applied by variety of molecular motors. Rather than being a rigid stick, DNA possesses dynamic structural variability that could be harnessed during critical steps of genome functioning. The strong relationship between DNA structure and key genomic processes necessitates the study of physical constrains acting on the double helix. Here we provide insight into the source, dynamics, and biology of DNA topological domains in the eukaryotic cells and summarize their possible involvement in gene transcription. We emphasize recent studies that might inspire and impact future experiments on the involvement of DNA topology in cellular functions.
Keywords: DNA topoisomerases; DNA topological domain; DNA topology; torsional stress; transcription.
Figures
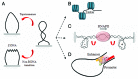
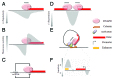
Comment on
- Kouzine F1, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517.
Similar articles
-
DNA torsion as a feedback mediator of transcription and chromatin dynamics.Nucleus. 2014 May-Jun;5(3):211-8. doi: 10.4161/nucl.29086. Epub 2014 May 12. Nucleus. 2014. PMID: 24819949 Free PMC article. Review.
-
DNA topoisomerase i as a transcription protein and a lethal cellular toxin.Ital J Biochem. 2007 Jun;56(2):122-9. Ital J Biochem. 2007. PMID: 17722653 Review.
-
The importance of being supercoiled: how DNA mechanics regulate dynamic processes.Biochim Biophys Acta. 2012 Jul;1819(7):632-8. doi: 10.1016/j.bbagrm.2011.12.007. Epub 2012 Jan 3. Biochim Biophys Acta. 2012. PMID: 22233557 Free PMC article. Review.
-
Histone and chromatin cross-talk.Curr Opin Cell Biol. 2003 Apr;15(2):172-83. doi: 10.1016/s0955-0674(03)00013-9. Curr Opin Cell Biol. 2003. PMID: 12648673 Review.
-
Novel orthogonal methods to uncover the complexity and diversity of nuclear architecture.Curr Opin Genet Dev. 2021 Apr;67:10-17. doi: 10.1016/j.gde.2020.10.002. Epub 2020 Nov 18. Curr Opin Genet Dev. 2021. PMID: 33220512 Review.
Cited by
-
Role of CTCF in DNA damage response.Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:61-68. doi: 10.1016/j.mrrev.2018.02.002. Epub 2018 Feb 23. Mutat Res Rev Mutat Res. 2019. PMID: 31395350 Free PMC article. Review.
-
Mechanical determinants of chromatin topology and gene expression.Nucleus. 2022 Dec;13(1):94-115. doi: 10.1080/19491034.2022.2038868. Nucleus. 2022. PMID: 35220881 Free PMC article. Review.
-
Protein/DNA interactions in complex DNA topologies: expect the unexpected.Biophys Rev. 2016;8(Suppl 1):145-155. doi: 10.1007/s12551-016-0241-7. Epub 2016 Nov 14. Biophys Rev. 2016. PMID: 28035245 Free PMC article. Review.
-
Topoisomerase IIα represses transcription by enforcing promoter-proximal pausing.Cell Rep. 2021 Apr 13;35(2):108977. doi: 10.1016/j.celrep.2021.108977. Cell Rep. 2021. PMID: 33852840 Free PMC article.
-
Loop-closure kinetics reveal a stable, right-handed DNA intermediate in Cre recombination.Nucleic Acids Res. 2020 May 7;48(8):4371-4381. doi: 10.1093/nar/gkaa153. Nucleic Acids Res. 2020. PMID: 32182357 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
