IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism
- PMID: 24755473
- PMCID: PMC4885639
- DOI: 10.1158/0008-5472.CAN-14-0772-T
IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism
Abstract
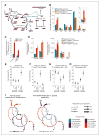
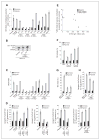
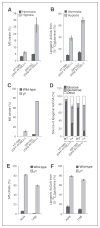
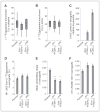
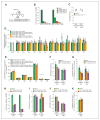
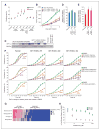
Oncogenic mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) occur in several types of cancer, but the metabolic consequences of these genetic changes are not fully understood. In this study, we performed (13)C metabolic flux analysis on a panel of isogenic cell lines containing heterozygous IDH1/2 mutations. We observed that under hypoxic conditions, IDH1-mutant cells exhibited increased oxidative tricarboxylic acid metabolism along with decreased reductive glutamine metabolism, but not IDH2-mutant cells. However, selective inhibition of mutant IDH1 enzyme function could not reverse the defect in reductive carboxylation activity. Furthermore, this metabolic reprogramming increased the sensitivity of IDH1-mutant cells to hypoxia or electron transport chain inhibition in vitro. Lastly, IDH1-mutant cells also grew poorly as subcutaneous xenografts within a hypoxic in vivo microenvironment. Together, our results suggest therapeutic opportunities to exploit the metabolic vulnerabilities specific to IDH1 mutation.
©2014 American Association for Cancer Research.
Conflict of interest statement
A.S. Divakaruni is a consultant/advisory board member of Seahorse Bioscience. M.G. Vander Heiden has ownership interest and is a consultant/advisory board member of Agios Pharmaceuticals. A.N. Murphy received a commercial research grant and is a consultant/advisory board member of Seahorse Bioscience. C. Straub, J.D. Growney, and R. Pagliarini have ownership interest in Novartis. C.M. Metallo has honoraria from the speakers’ bureau of Agios Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
Figures

Similar articles
-
Reductive carboxylation supports redox homeostasis during anchorage-independent growth.Nature. 2016 Apr 14;532(7598):255-8. doi: 10.1038/nature17393. Epub 2016 Apr 6. Nature. 2016. PMID: 27049945 Free PMC article.
-
IDH2 compensates for IDH1 mutation to maintain cell survival under hypoxic conditions in IDH1‑mutant tumor cells.Mol Med Rep. 2019 Aug;20(2):1893-1900. doi: 10.3892/mmr.2019.10418. Epub 2019 Jun 24. Mol Med Rep. 2019. PMID: 31257503
-
Cancer-associated isocitrate dehydrogenase 1 (IDH1) R132H mutation and d-2-hydroxyglutarate stimulate glutamine metabolism under hypoxia.J Biol Chem. 2014 Aug 22;289(34):23318-28. doi: 10.1074/jbc.M114.575183. Epub 2014 Jul 1. J Biol Chem. 2014. PMID: 24986863 Free PMC article.
-
Development of Novel Therapeutics Targeting Isocitrate Dehydrogenase Mutations in Cancer.Curr Top Med Chem. 2018;18(6):505-524. doi: 10.2174/1568026618666180518091144. Curr Top Med Chem. 2018. PMID: 29773061 Review.
-
Metabolic consequences of oncogenic IDH mutations.Pharmacol Ther. 2015 Aug;152:54-62. doi: 10.1016/j.pharmthera.2015.05.003. Epub 2015 May 5. Pharmacol Ther. 2015. PMID: 25956465 Free PMC article. Review.
Cited by
-
Hyperpolarized (13)C MR imaging detects no lactate production in mutant IDH1 gliomas: Implications for diagnosis and response monitoring.Neuroimage Clin. 2016 Jun 23;12:180-9. doi: 10.1016/j.nicl.2016.06.018. eCollection 2016. Neuroimage Clin. 2016. PMID: 27437179 Free PMC article.
-
Altered Metabolism in Glioblastoma: Myeloid-Derived Suppressor Cell (MDSC) Fitness and Tumor-Infiltrating Lymphocyte (TIL) Dysfunction.Int J Mol Sci. 2021 Apr 24;22(9):4460. doi: 10.3390/ijms22094460. Int J Mol Sci. 2021. PMID: 33923299 Free PMC article. Review.
-
Differential metabolic alterations in IDH1 mutant vs. wildtype glioma cells promote epileptogenesis through distinctive mechanisms.Front Cell Neurosci. 2023 Nov 9;17:1288918. doi: 10.3389/fncel.2023.1288918. eCollection 2023. Front Cell Neurosci. 2023. PMID: 38026690 Free PMC article. Review.
-
IDH1-mutant preleukemic hematopoietic stem cells can be eliminated by inhibition of oxidative phosphorylation.Blood Cancer Discov. 2023 Dec 13;5(2):114-31. doi: 10.1158/2643-3230.BCD-23-0195. Online ahead of print. Blood Cancer Discov. 2023. PMID: 38091010 Free PMC article.
-
Targeting mitochondrial respiration for the treatment of acute myeloid leukemia.Biochem Pharmacol. 2020 Dec;182:114253. doi: 10.1016/j.bcp.2020.114253. Epub 2020 Oct 2. Biochem Pharmacol. 2020. PMID: 33011159 Free PMC article. Review.
References
-
- Arai M, Nobusawa S, Ikota H, Takemura S, Nakazato Y. Frequent IDH1/2 mutations in intracranial chondrosarcoma: a possible diagnostic clue for its differentiation from chordoma. Brain Tumor Pathol. 2012;29:201–6. - PubMed
-
- Hayden JT, Fruhwald MC, Hasselblatt M, Ellison DW, Bailey S, Clifford SC. Frequent IDH1 mutations in supratentorial primitive neuroectodermal tumors (sPNET) of adults but not children. Cell Cycle. 2009;8:1806–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

