A conserved hydrogen-bonding network of P2 bis-tetrahydrofuran-containing HIV-1 protease inhibitors (PIs) with a protease active-site amino acid backbone aids in their activity against PI-resistant HIV
- PMID: 24752271
- PMCID: PMC4068604
- DOI: 10.1128/AAC.00107-14
A conserved hydrogen-bonding network of P2 bis-tetrahydrofuran-containing HIV-1 protease inhibitors (PIs) with a protease active-site amino acid backbone aids in their activity against PI-resistant HIV
Abstract
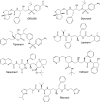
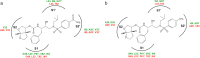
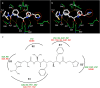
In the present study, GRL008, a novel nonpeptidic human immunodeficiency virus type 1 (HIV-1) protease inhibitor (PI), and darunavir (DRV), both of which contain a P2-bis-tetrahydrofuranyl urethane (bis-THF) moiety, were found to exert potent antiviral activity (50% effective concentrations [EC50s], 0.029 and 0.002 μM, respectively) against a multidrug-resistant clinical isolate of HIV-1 (HIVA02) compared to ritonavir (RTV; EC50, >1.0 μM) and tipranavir (TPV; EC50, 0.364 μM). Additionally, GRL008 showed potent antiviral activity against an HIV-1 variant selected in the presence of DRV over 20 passages (HIVDRV(R)P20), with a 2.6-fold increase in its EC50 (0.097 μM) compared to its corresponding EC50 (0.038 μM) against wild-type HIV-1NL4-3 (HIVWT). Based on X-ray crystallographic analysis, both GRL008 and DRV showed strong hydrogen bonds (H-bonds) with the backbone-amide nitrogen/carbonyl oxygen atoms of conserved active-site amino acids G27, D29, D30, and D30' of HIVA02 protease (PRA02) and wild-type PR in their corresponding crystal structures, while TPV lacked H-bonds with G27 and D30' due to an absence of polar groups. The P2' thiazolyl moiety of RTV showed two conformations in the crystal structure of the PRA02-RTV complex, one of which showed loss of contacts in the S2' binding pocket of PRA02, supporting RTV's compromised antiviral activity (EC50, >1 μM). Thus, the conserved H-bonding network of P2-bis-THF-containing GRL008 with the backbone of G27, D29, D30, and D30' most likely contributes to its persistently greater antiviral activity against HIVWT, HIVA02, and HIVDRV(R)P20.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
GRL-0519, a novel oxatricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor (PI), potently suppresses replication of a wide spectrum of multi-PI-resistant HIV-1 variants in vitro.Antimicrob Agents Chemother. 2013 May;57(5):2036-46. doi: 10.1128/AAC.02189-12. Epub 2013 Feb 12. Antimicrob Agents Chemother. 2013. PMID: 23403426 Free PMC article.
-
GRL-079, a Novel HIV-1 Protease Inhibitor, Is Extremely Potent against Multidrug-Resistant HIV-1 Variants and Has a High Genetic Barrier against the Emergence of Resistant Variants.Antimicrob Agents Chemother. 2018 Apr 26;62(5):e02060-17. doi: 10.1128/AAC.02060-17. Print 2018 May. Antimicrob Agents Chemother. 2018. PMID: 29463535 Free PMC article.
-
Novel Protease Inhibitors Containing C-5-Modified bis-Tetrahydrofuranylurethane and Aminobenzothiazole as P2 and P2' Ligands That Exert Potent Antiviral Activity against Highly Multidrug-Resistant HIV-1 with a High Genetic Barrier against the Emergence of Drug Resistance.Antimicrob Agents Chemother. 2019 Jul 25;63(8):e00372-19. doi: 10.1128/AAC.00372-19. Print 2019 Aug. Antimicrob Agents Chemother. 2019. PMID: 31085520 Free PMC article.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Tipranavir: a protease inhibitor from a new class with distinct antiviral activity.J Acquir Immune Defic Syndr. 2003 Sep;34 Suppl 1:S91-4. doi: 10.1097/00126334-200309011-00014. J Acquir Immune Defic Syndr. 2003. PMID: 14562864 Review.
Cited by
-
Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.ACS Infect Dis. 2019 Feb 8;5(2):316-325. doi: 10.1021/acsinfecdis.8b00336. Epub 2018 Dec 31. ACS Infect Dis. 2019. PMID: 30543749 Free PMC article.
-
Aureonitol, a Fungi-Derived Tetrahydrofuran, Inhibits Influenza Replication by Targeting Its Surface Glycoprotein Hemagglutinin.PLoS One. 2015 Oct 13;10(10):e0139236. doi: 10.1371/journal.pone.0139236. eCollection 2015. PLoS One. 2015. PMID: 26462111 Free PMC article.
-
Design, Synthesis, and Biological Evaluation of Darunavir Analogs as HIV-1 Protease Inhibitors.ACS Bio Med Chem Au. 2024 Sep 19;4(5):242-256. doi: 10.1021/acsbiomedchemau.4c00040. eCollection 2024 Oct 16. ACS Bio Med Chem Au. 2024. PMID: 39431267 Free PMC article.
-
Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.J Biol Chem. 2016 Oct 21;291(43):22741-22756. doi: 10.1074/jbc.M116.747568. Epub 2016 Aug 30. J Biol Chem. 2016. PMID: 27576689 Free PMC article.
-
Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS.J Med Chem. 2016 Jun 9;59(11):5172-208. doi: 10.1021/acs.jmedchem.5b01697. Epub 2016 Jan 22. J Med Chem. 2016. PMID: 26799988 Free PMC article. Review.
References
-
- Yedidi RS, Proteasa G, Martinez JL, Vickrey JF, Martin PD, Wawrzak Z, Liu Z, Kovari IA, Kovari LC. 2011. Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants. Acta Crystallogr. D Biol. Crystallogr. 67:524–532. 10.1107/S0907444911011541 - DOI - PMC - PubMed
-
- Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang Y-F, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. 2003. Novel bis-tetrahydrofuranylurethan-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 47:3123–3129. 10.1128/AAC.47.10.3123-3129.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

