The membrane proteome of sensory cilia to the depth of olfactory receptors
- PMID: 24748648
- PMCID: PMC4083118
- DOI: 10.1074/mcp.M113.035378
The membrane proteome of sensory cilia to the depth of olfactory receptors
Abstract
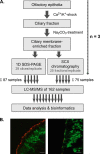
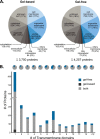
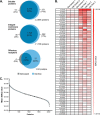
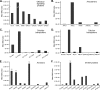
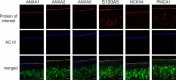
In the nasal cavity, the nonmotile cilium of olfactory sensory neurons (OSNs) constitutes the chemosensory interface between the ambient environment and the brain. The unique sensory organelle facilitates odor detection for which it includes all necessary components of initial and downstream olfactory signal transduction. In addition to its function in olfaction, a more universal role in modulating different signaling pathways is implicated, for example, in neurogenesis, apoptosis, and neural regeneration. To further extend our knowledge about this multifunctional signaling organelle, it is of high importance to establish a most detailed proteome map of the ciliary membrane compartment down to the level of transmembrane receptors. We detached cilia from mouse olfactory epithelia via Ca(2+)/K(+) shock followed by the enrichment of ciliary membrane proteins at alkaline pH, and we identified a total of 4,403 proteins by gel-based and gel-free methods in conjunction with high resolution LC/MS. This study is the first to report the detection of 62 native olfactory receptor proteins and to provide evidence for their heterogeneous expression at the protein level. Quantitative data evaluation revealed four ciliary membrane-associated candidate proteins (the annexins ANXA1, ANXA2, ANXA5, and S100A5) with a suggested function in the regulation of olfactory signal transduction, and their presence in ciliary structures was confirmed by immunohistochemistry. Moreover, we corroborated the ciliary localization of the potassium-dependent Na(+)/Ca(2+) exchanger (NCKX) 4 and the plasma membrane Ca(2+)-ATPase 1 (PMCA1) involved in olfactory signal termination, and we detected for the first time NCKX2 in olfactory cilia. Through comparison with transcriptome data specific for mature, ciliated OSNs, we finally delineated the membrane ciliome of OSNs. The membrane proteome of olfactory cilia established here is the most complete today, thus allowing us to pave new avenues for the study of diverse molecular functions and signaling pathways in and out of olfactory cilia and thus to advance our understanding of the biology of sensory organelles in general.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
The proteome of rat olfactory sensory cilia.Proteomics. 2009 Jan;9(2):322-34. doi: 10.1002/pmic.200800149. Proteomics. 2009. PMID: 19086097
-
Plasma membrane Ca(2+)-ATPase in the cilia of olfactory receptor neurons: possible role in Ca(2+) clearance.Eur J Neurosci. 2007 Nov;26(9):2524-31. doi: 10.1111/j.1460-9568.2007.05863.x. Epub 2007 Oct 23. Eur J Neurosci. 2007. PMID: 17970729
-
Molecular components of signal amplification in olfactory sensory cilia.Proc Natl Acad Sci U S A. 2010 Mar 30;107(13):6052-7. doi: 10.1073/pnas.0909032107. Epub 2010 Mar 15. Proc Natl Acad Sci U S A. 2010. PMID: 20231443 Free PMC article.
-
Maturation of the Olfactory Sensory Neuron and Its Cilia.Chem Senses. 2020 Dec 5;45(9):805-822. doi: 10.1093/chemse/bjaa070. Chem Senses. 2020. PMID: 33075817 Free PMC article. Review.
-
Engineering Aspects of Olfaction.In: Persaud KC, Marco S, Gutiérrez-Gálvez A, editors. Neuromorphic Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2013. Chapter 1. In: Persaud KC, Marco S, Gutiérrez-Gálvez A, editors. Neuromorphic Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2013. Chapter 1. PMID: 26042329 Free Books & Documents. Review.
Cited by
-
Astrogliosis and neuroinflammation underlie scoliosis upon cilia dysfunction.Elife. 2024 Oct 10;13:RP96831. doi: 10.7554/eLife.96831. Elife. 2024. PMID: 39388365 Free PMC article.
-
Energy Requirements of Odor Transduction in the Chemosensory Cilia of Olfactory Sensory Neurons Rely on Oxidative Phosphorylation and Glycolytic Processing of Extracellular Glucose.J Neurosci. 2017 Jun 7;37(23):5736-5743. doi: 10.1523/JNEUROSCI.2640-16.2017. Epub 2017 May 12. J Neurosci. 2017. PMID: 28500222 Free PMC article.
-
Endogenous zinc nanoparticles in the rat olfactory epithelium are functionally significant.Sci Rep. 2020 Oct 28;10(1):18435. doi: 10.1038/s41598-020-75430-w. Sci Rep. 2020. PMID: 33116197 Free PMC article.
-
Mixture interactions at mammalian olfactory receptors are dependent on the cellular environment.Sci Rep. 2021 Apr 29;11(1):9278. doi: 10.1038/s41598-021-88601-0. Sci Rep. 2021. PMID: 33927269 Free PMC article.
-
Identification and Characterization of Proteins That Are Involved in RTP1S-Dependent Transport of Olfactory Receptors.Int J Mol Sci. 2023 Apr 25;24(9):7829. doi: 10.3390/ijms24097829. Int J Mol Sci. 2023. PMID: 37175532 Free PMC article.
References
-
- McEwen D. P., Jenkins P. M., Martens J. R. (2008) Olfactory cilia: our direct neuronal connection to the external world. Curr. Top. Dev. Biol. 85, 333–370 - PubMed
-
- Buck L., Axel R. (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 - PubMed
-
- Breer H. (2003) Sense of smell: recognition and transduction of olfactory signals. Biochem. Soc. Trans 31, 113–116 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

