The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo
- PMID: 24737863
- PMCID: PMC3983907
- DOI: 10.7554/eLife.01861
The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo
Abstract
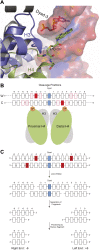
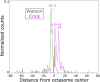
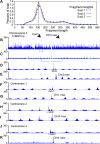
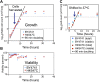
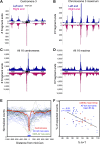
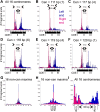
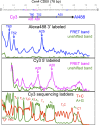
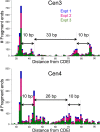
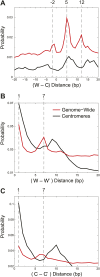
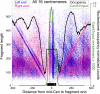
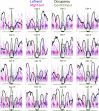
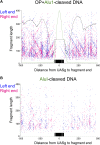
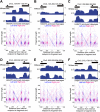
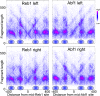
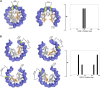
In budding yeast, a single cenH3 (Cse4) nucleosome occupies the ∼120-bp functional centromere, however conflicting structural models for the particle have been proposed. To resolve this controversy, we have applied H4S47C-anchored cleavage mapping, which reveals the precise position of histone H4 in every nucleosome in the genome. We find that cleavage patterns at centromeres are unique within the genome and are incompatible with symmetrical structures, including octameric nucleosomes and (Cse4/H4)2 tetrasomes. Centromere cleavage patterns are compatible with a precisely positioned core structure, one in which each of the 16 yeast centromeres is occupied by oppositely oriented Cse4/H4/H2A/H2B hemisomes in two rotational phases within the population. Centromere-specific hemisomes are also inferred from distances observed between closely-spaced H4 cleavages, as predicted from structural modeling. Our results indicate that the orientation and rotational position of the stable hemisome at each yeast centromere is not specified by the functional centromere sequence. DOI: http://dx.doi.org/10.7554/eLife.01861.001.
Keywords: centromeres; chemical cleavage mapping; nucleosome.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

Similar articles
-
Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast.Mol Cell. 2011 Aug 5;43(3):369-80. doi: 10.1016/j.molcel.2011.07.009. Mol Cell. 2011. PMID: 21816344 Free PMC article.
-
Reconstitution of hemisomes on budding yeast centromeric DNA.Nucleic Acids Res. 2013 Jun;41(11):5769-83. doi: 10.1093/nar/gkt314. Epub 2013 Apr 24. Nucleic Acids Res. 2013. PMID: 23620291 Free PMC article.
-
Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.Nature. 2011 Apr 14;472(7342):234-7. doi: 10.1038/nature09854. Epub 2011 Mar 16. Nature. 2011. PMID: 21412236 Free PMC article.
-
The unconventional structure of centromeric nucleosomes.Chromosoma. 2012 Aug;121(4):341-52. doi: 10.1007/s00412-012-0372-y. Epub 2012 May 3. Chromosoma. 2012. PMID: 22552438 Free PMC article. Review.
-
Structure, dynamics, and evolution of centromeric nucleosomes.Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):15974-81. doi: 10.1073/pnas.0707648104. Epub 2007 Sep 24. Proc Natl Acad Sci U S A. 2007. PMID: 17893333 Free PMC article. Review.
Cited by
-
Centromeres under Pressure: Evolutionary Innovation in Conflict with Conserved Function.Genes (Basel). 2020 Aug 10;11(8):912. doi: 10.3390/genes11080912. Genes (Basel). 2020. PMID: 32784998 Free PMC article. Review.
-
Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.Mol Cell. 2024 Sep 19;84(18):3423-3437.e8. doi: 10.1016/j.molcel.2024.08.022. Epub 2024 Sep 12. Mol Cell. 2024. PMID: 39270644
-
Transcription dynamically patterns the meiotic chromosome-axis interface.Elife. 2015 Aug 10;4:e07424. doi: 10.7554/eLife.07424. Elife. 2015. PMID: 26258962 Free PMC article.
-
Reb1, Cbf1, and Pho4 Bias Histone Sliding and Deposition Away from Their Binding Sites.Mol Cell Biol. 2022 Feb 17;42(2):e0047221. doi: 10.1128/MCB.00472-21. Epub 2021 Dec 13. Mol Cell Biol. 2022. PMID: 34898278 Free PMC article.
-
An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites.Elife. 2017 Jan 16;6:e21856. doi: 10.7554/eLife.21856. Elife. 2017. PMID: 28079019 Free PMC article.
References
-
- Bartfai R, Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Janssen-Megens E, Kaan A, Treeck M, Gilberger TW, Francoijs KJ, Stunnenberg HG. 2010. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLOS Pathogens 6:e1001223. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

