The power of neuroimaging biomarkers for screening frontotemporal dementia
- PMID: 24687814
- PMCID: PMC4107021
- DOI: 10.1002/hbm.22515
The power of neuroimaging biomarkers for screening frontotemporal dementia
Abstract
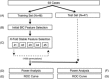
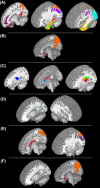
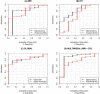
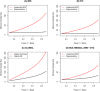
Frontotemporal dementia (FTD) is a clinically and pathologically heterogeneous neurodegenerative disease that can result from either frontotemporal lobar degeneration (FTLD) or Alzheimer's disease (AD) pathology. It is critical to establish statistically powerful biomarkers that can achieve substantial cost-savings and increase the feasibility of clinical trials. We assessed three broad categories of neuroimaging methods to screen underlying FTLD and AD pathology in a clinical FTD series: global measures (e.g., ventricular volume), anatomical volumes of interest (VOIs) (e.g., hippocampus) using a standard atlas, and data-driven VOIs using Eigenanatomy. We evaluated clinical FTD patients (N = 93) with cerebrospinal fluid, gray matter (GM) magnetic resonance imaging (MRI), and diffusion tensor imaging (DTI) to assess whether they had underlying FTLD or AD pathology. Linear regression was performed to identify the optimal VOIs for each method in a training dataset and then we evaluated classification sensitivity and specificity in an independent test cohort. Power was evaluated by calculating minimum sample sizes required in the test classification analyses for each model. The data-driven VOI analysis using a multimodal combination of GM MRI and DTI achieved the greatest classification accuracy (89% sensitive and 89% specific) and required a lower minimum sample size (N = 26) relative to anatomical VOI and global measures. We conclude that a data-driven VOI approach using Eigenanatomy provides more accurate classification, benefits from increased statistical power in unseen datasets, and therefore provides a robust method for screening underlying pathology in FTD patients for entry into clinical trials.
Keywords: Alzheimer's disease; DTI; MRI; biomarkers; classification; frontotemporal degeneration; statistical power.
Copyright © 2014 Wiley Periodicals, Inc.
Figures
Similar articles
-
Contribution of CSF biomarkers to early-onset Alzheimer's disease and frontotemporal dementia neuroimaging signatures.Hum Brain Mapp. 2020 Jun 1;41(8):2004-2013. doi: 10.1002/hbm.24925. Epub 2020 Jan 16. Hum Brain Mapp. 2020. PMID: 31944489 Free PMC article.
-
White matter imaging helps dissociate tau from TDP-43 in frontotemporal lobar degeneration.J Neurol Neurosurg Psychiatry. 2013 Sep;84(9):949-55. doi: 10.1136/jnnp-2012-304418. Epub 2013 Mar 9. J Neurol Neurosurg Psychiatry. 2013. PMID: 23475817 Free PMC article.
-
Cerebrospinal Fluid Biomarkers in Patients with Frontotemporal Dementia Spectrum: A Single-Center Study.J Alzheimers Dis. 2018;66(2):551-563. doi: 10.3233/JAD-180409. J Alzheimers Dis. 2018. PMID: 30320576
-
Cerebrospinal Fluid Biomarkers for the Differential Diagnosis between Alzheimer's Disease and Frontotemporal Lobar Degeneration: Systematic Review, HSROC Analysis, and Confounding Factors.J Alzheimers Dis. 2017;55(2):625-644. doi: 10.3233/JAD-160366. J Alzheimers Dis. 2017. PMID: 27716663 Review.
-
Blood Biomarkers for the Diagnosis of Neurodegenerative Dementia: A Systematic Review.J Geriatr Psychiatry Neurol. 2023 Jul;36(4):267-281. doi: 10.1177/08919887221141651. Epub 2022 Nov 24. J Geriatr Psychiatry Neurol. 2023. PMID: 36423207 Review.
Cited by
-
Brain network efficiency is influenced by the pathologic source of corticobasal syndrome.Neurology. 2017 Sep 26;89(13):1373-1381. doi: 10.1212/WNL.0000000000004324. Epub 2017 Aug 4. Neurology. 2017. PMID: 28779011 Free PMC article.
-
Atrophy in behavioural variant frontotemporal dementia spans multiple large-scale prefrontal and temporal networks.Brain. 2023 Nov 2;146(11):4476-4485. doi: 10.1093/brain/awad167. Brain. 2023. PMID: 37201288 Free PMC article.
-
Neurodegenerative disease: MRI biomarkers - a precision medicine tool in neurology?Nat Rev Neurol. 2016 Jun;12(6):323-4. doi: 10.1038/nrneurol.2016.51. Epub 2016 Apr 15. Nat Rev Neurol. 2016. PMID: 27080517 Free PMC article.
-
Morphometric MRI as a diagnostic biomarker of frontotemporal dementia: A systematic review to determine clinical applicability.Neuroimage Clin. 2018;20:685-696. doi: 10.1016/j.nicl.2018.08.028. Epub 2018 Aug 31. Neuroimage Clin. 2018. PMID: 30218900 Free PMC article.
-
Measurement Challenges in Research With Individuals With Cognitive Impairment.Res Gerontol Nurs. 2019 Jan 1;12(1):7-15. doi: 10.3928/19404921-20181212-06. Res Gerontol Nurs. 2019. PMID: 30653646 Free PMC article.
References
-
- Alexander DC, Pierpaoli C, Basser PJ, Gee JC (2001): Spatial transformations of diffusion tensor magnetic resonance images. IEEE Trans Med Imaging 20:1131–1139. - PubMed
-
- Armstrong MJ, Litvan I, Lang AE, Bak TH, Bhatia KP, Borroni B, Boxer AL, Dickson DW, Grossman M, Hallett M, Josephs KA, Kertesz A, Lee SE, Miller BL, Reich SG, Riley DE, Tolosa E, Tröster AI, Vidailhet M, Weiner WJ (2013): Criteria for the diagnosis of corticobasal degeneration. Neurology 80:496–503. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

