eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining
- PMID: 24686316
- PMCID: PMC4193923
- DOI: 10.1002/embj.201387344
eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining
Abstract
eIF5B is a eukaryal translational GTPase that catalyzes ribosomal subunit joining to form elongation-competent ribosomes. Despite its central role in protein synthesis, the mechanistic details that govern the function of eIF5B or its archaeal and bacterial (IF2) orthologs remained unclear. Here, we present six high-resolution crystal structures of eIF5B in its apo, GDP- and GTP-bound form that, together with an analysis of the thermodynamics of nucleotide binding, provide a detailed picture of the entire nucleotide cycle performed by eIF5B. Our data show that GTP binding induces significant conformational changes in the two conserved switch regions of the G domain, resulting in the reorganization of the GTPase center. These rearrangements are accompanied by the rotation of domain II relative to the G domain and release of domain III from its stable contacts with switch 2, causing an increased intrinsic flexibility in the free GTP-bound eIF5B. Based on these data, we propose a novel domain release mechanism for eIF5B/IF2 activation that explains how eIF5B and IF2 fulfill their catalytic role during ribosomal subunit joining.
Keywords: GTPase; crystal structure; molecular machines; ribosome; subunit joining; translation initiation.
© 2014 The Authors.
Figures
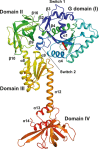
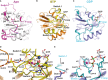
G domain-based superposition of domains I (G), II and III from Ct-eIF5B in the apo and GTP-bound state. Both G domains are shown in gray; otherwise the same color code was used as in Fig2. The GTP-induced rearrangements result in the loss of interactions between switch 2 and helices α8 and α9 (circle) and ultimately in the release of domain III from the G domain. Domain II rotates by ∼30° relative to the G domain and is stabilized in its new orientation by the newly formed contact between β13 and β14 loop and domain I.
In apo eIF5B, the inactive switch 2 (pink) forms stable contacts with helices α8 and α9 of domain III which are broken upon the GTP-induced transition of switch 2 to its active state (yellow).
Comparison of the molecular switch mechanisms in EF-Tu (left) and eIF5B (right). Both trGTPases are shown in their inactive apo or GDP-bound (top) and GTP-bound (bottom) states, respectively. Functionally relevant interactions between the switch regions (yellow) of the G domain and downstream functional domains (blue) are indicated by dashed circles.
The contact surface (pink) found between domain III (blue) and domains I and II in apo eIF5B (top) is entirely lost in ribosome-bound eIF5B·GDPCP (bottom; PDB: 4BVX (Fernandez et al, 2013)), where domains III and IV become stabilized between SRL and Met-tRNAiMet (not shown).

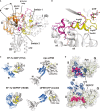
Superposition of the catalytic centers of eIF5B·GTP (yellow) and free EF-Tu·GDPNP (cyan; PDB: 1EXM). Conserved residues are shown as sticks; GDPNP is omitted for clarity.
Superposition of the catalytic centers of eIF5B·GTP (yellow) and ribosome-bound EF-Tu·GDPCP (PDB: 2XQD, 2XQE) with the sarcin-ricin loop (SRL) in pink. Structural alterations relative to free eIF5B·GTP and EF-Tu·GDPNP are limited to Hiscat of EF-Tu, which is reoriented (arrow) into its active position between A2662 and Wcat.
Model of domains I and II of eIF5B (orange) on the ribosome, based on the superposition with EF-Tu·GDPCP. Similar to eIF5B·GDPCP in the cryo-EM model of the 80S IC (see Supplementary Fig S5B), the G domain is associated with the SRL of the large subunit (LSU; green), while domain II interacts with the body of the small subunit (SSU; light pink).
Putative interactions between the G domain and the SRL/H95 (green). Direct interactions are indicated by black dashed lines; red dashed lines indicate the positions in H95 that are cleaved by Fe(II)-BABE introduced in the position of Lys540 (Unbehaun et al, 2007). His505 lies only 3.5 Å from H95, explaining why the H505Y mutation results in a reduced affinity for the ribosome and GTPase deficiency in eIF5B (Shin et al, 2002) (see also Supplementary Table S1). The conserved Arg534 likely contributes to the interactions with H95.

Heat capacity changes upon eIF5B interaction with GDP or GTP. Temperature dependency of binding enthalpy changes (ΔH) upon Ct-eIF5B(517C) interactions with GDP in the presence (•) or absence (○) of MgCl2 and of Ct-eIF5B(517C) (▾) and Ct-eIF5B(517–852) (Δ) with GTP in the presence of MgCl2. Standard deviations are given by error bars (in some cases not visible because they are smaller than the symbol size).
Domains I–III of apo Ct-eIF5B. Indicated are the contact areas of domain III to domains I and II, respectively.
Similar articles
-
Intragenic suppressor mutations restore GTPase and translation functions of a eukaryotic initiation factor 5B switch II mutant.Mol Cell Biol. 2007 Mar;27(5):1677-85. doi: 10.1128/MCB.01258-06. Epub 2006 Dec 22. Mol Cell Biol. 2007. PMID: 17189426 Free PMC article.
-
Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining.Mol Cell Biol. 2007 Mar;27(6):2384-97. doi: 10.1128/MCB.02254-06. Epub 2007 Jan 22. Mol Cell Biol. 2007. PMID: 17242201 Free PMC article.
-
Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining.J Biol Chem. 2006 Mar 31;281(13):8469-75. doi: 10.1074/jbc.M600210200. Epub 2006 Feb 3. J Biol Chem. 2006. PMID: 16461768
-
Assembling the archaeal ribosome: roles for translation-factor-related GTPases.Biochem Soc Trans. 2011 Jan;39(1):45-50. doi: 10.1042/BST0390045. Biochem Soc Trans. 2011. PMID: 21265745 Review.
-
Mechanism of ribosomal subunit joining during eukaryotic translation initiation.Biochem Soc Trans. 2008 Aug;36(Pt 4):653-7. doi: 10.1042/BST0360653. Biochem Soc Trans. 2008. PMID: 18631135 Review.
Cited by
-
Saccharomyces cerevisiae Ski7 Is a GTP-Binding Protein Adopting the Characteristic Conformation of Active Translational GTPases.Structure. 2015 Jul 7;23(7):1336-43. doi: 10.1016/j.str.2015.04.018. Epub 2015 Jun 4. Structure. 2015. PMID: 26051716 Free PMC article.
-
Characterizing Cellular Responses During Oncolytic Maraba Virus Infection.Int J Mol Sci. 2019 Jan 29;20(3):580. doi: 10.3390/ijms20030580. Int J Mol Sci. 2019. PMID: 30700020 Free PMC article.
-
Ribosome-induced tuning of GTP hydrolysis by a translational GTPase.Proc Natl Acad Sci U S A. 2014 Oct 7;111(40):14418-23. doi: 10.1073/pnas.1412676111. Epub 2014 Sep 22. Proc Natl Acad Sci U S A. 2014. PMID: 25246550 Free PMC article.
-
eIF5B increases ASAP1 expression to promote HCC proliferation and invasion.Oncotarget. 2016 Sep 20;7(38):62327-62339. doi: 10.18632/oncotarget.11469. Oncotarget. 2016. PMID: 27694689 Free PMC article.
-
Structural basis for the transition from translation initiation to elongation by an 80S-eIF5B complex.Nat Commun. 2020 Oct 6;11(1):5003. doi: 10.1038/s41467-020-18829-3. Nat Commun. 2020. PMID: 33024099 Free PMC article.
References
-
- Abrahamson JK, Laue TM, Miller DL, Johnson AE. Direct determination of the association constant between elongation factor Tu X GTP and aminoacyl-tRNA using fluorescence. Biochemistry. 1985;24:692–700. - PubMed
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. - PMC - PubMed
-
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

