The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor
- PMID: 24663151
- PMCID: PMC3963952
- DOI: 10.1371/journal.pone.0092727
The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor
Abstract
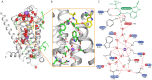
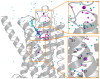
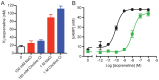
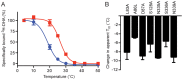
The β1-adrenoceptor (β1AR) is a G protein-coupled receptor (GPCR) that is activated by the endogenous agonists adrenaline and noradrenaline. We have determined the structure of an ultra-thermostable β1AR mutant bound to the weak partial agonist cyanopindolol to 2.1 Å resolution. High-quality crystals (100 μm plates) were grown in lipidic cubic phase without the assistance of a T4 lysozyme or BRIL fusion in cytoplasmic loop 3, which is commonly employed for GPCR crystallisation. An intramembrane Na+ ion was identified co-ordinated to Asp872.50, Ser1283.39 and 3 water molecules, which is part of a more extensive network of water molecules in a cavity formed between transmembrane helices 1, 2, 3, 6 and 7. Remarkably, this water network and Na+ ion is highly conserved between β1AR and the adenosine A2A receptor (rmsd of 0.3 Å), despite an overall rmsd of 2.4 Å for all Cα atoms and only 23% amino acid identity in the transmembrane regions. The affinity of agonist binding and nanobody Nb80 binding to β1AR is unaffected by Na+ ions, but the stability of the receptor is decreased by 7.5°C in the absence of Na+. Mutation of amino acid side chains that are involved in the co-ordination of either Na+ or water molecules in the network decreases the stability of β1AR by 5-10°C. The data suggest that the intramembrane Na+ and associated water network stabilise the ligand-free state of β1AR, but still permits the receptor to form the activated state which involves the collapse of the Na+ binding pocket on agonist binding.
Conflict of interest statement
Figures
Similar articles
-
Structure of a beta1-adrenergic G-protein-coupled receptor.Nature. 2008 Jul 24;454(7203):486-91. doi: 10.1038/nature07101. Epub 2008 Jun 25. Nature. 2008. PMID: 18594507 Free PMC article.
-
Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound β1-Adrenergic Receptor.Mol Pharmacol. 2015 Dec;88(6):1024-34. doi: 10.1124/mol.115.101030. Epub 2015 Sep 18. Mol Pharmacol. 2015. PMID: 26385885 Free PMC article.
-
Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor.Nature. 2016 Feb 11;530(7589):237-41. doi: 10.1038/nature16577. Epub 2016 Feb 3. Nature. 2016. PMID: 26840483
-
Agonist-bound structures of G protein-coupled receptors.Curr Opin Struct Biol. 2012 Aug;22(4):482-90. doi: 10.1016/j.sbi.2012.03.007. Epub 2012 Apr 3. Curr Opin Struct Biol. 2012. PMID: 22480933 Review.
-
Domains of beta1 and beta2 adrenergic receptors to bind subtype selective agonists.Life Sci. 1998;62(17-18):1513-7. doi: 10.1016/s0024-3205(98)00099-x. Life Sci. 1998. PMID: 9585128 Review.
Cited by
-
G Protein-Coupled Receptor-Ligand Pose and Functional Class Prediction.Int J Mol Sci. 2024 Jun 22;25(13):6876. doi: 10.3390/ijms25136876. Int J Mol Sci. 2024. PMID: 38999982 Free PMC article.
-
Membrane potentials regulating GPCRs: insights from experiments and molecular dynamics simulations.Curr Opin Pharmacol. 2016 Oct;30:44-50. doi: 10.1016/j.coph.2016.06.011. Epub 2016 Jul 27. Curr Opin Pharmacol. 2016. PMID: 27474871 Free PMC article. Review.
-
G protein-coupled receptors of class A harness the energy of membrane potential to increase their sensitivity and selectivity.Biochim Biophys Acta Biomembr. 2019 Dec 1;1861(12):183051. doi: 10.1016/j.bbamem.2019.183051. Epub 2019 Aug 23. Biochim Biophys Acta Biomembr. 2019. PMID: 31449800 Free PMC article.
-
Molecular dynamics-guided discovery of an ago-allosteric modulator for GPR40/FFAR1.Proc Natl Acad Sci U S A. 2019 Apr 2;116(14):7123-7128. doi: 10.1073/pnas.1811066116. Epub 2019 Mar 14. Proc Natl Acad Sci U S A. 2019. PMID: 30872479 Free PMC article.
-
The action of a negative allosteric modulator at the dopamine D2 receptor is dependent upon sodium ions.Sci Rep. 2018 Jan 19;8(1):1208. doi: 10.1038/s41598-018-19642-1. Sci Rep. 2018. PMID: 29352161 Free PMC article.
References
-
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, et al. (2013) Molecular signatures of G-protein-coupled receptors. Nature 494: 185–194. - PubMed
-
- Tate CG, Schertler GF (2009) Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol 19: 386–395. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

