Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N-glycolyl neuraminic acid as a receptor candidate
- PMID: 24648448
- PMCID: PMC4093890
- DOI: 10.1128/JVI.03455-13
Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N-glycolyl neuraminic acid as a receptor candidate
Abstract
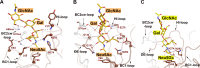
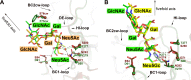
Human polyomavirus 9 (HPyV9) is a closely related homologue of simian B-lymphotropic polyomavirus (LPyV). In order to define the architecture and receptor binding properties of HPyV9, we solved high-resolution crystal structures of its major capsid protein, VP1, in complex with three putative oligosaccharide receptors identified by glycan microarray screening. Comparison of the properties of HPyV9 VP1 with the known structure and glycan-binding properties of LPyV VP1 revealed that both viruses engage short sialylated oligosaccharides, but small yet important differences in specificity were detected. Surprisingly, HPyV9 VP1 preferentially binds sialyllactosamine compounds terminating in 5-N-glycolyl neuraminic acid (Neu5Gc) over those terminating in 5-N-acetyl neuraminic acid (Neu5Ac), whereas LPyV does not exhibit such a preference. The structural analysis demonstrated that HPyV9 makes specific contacts, via hydrogen bonds, with the extra hydroxyl group present in Neu5Gc. An equivalent hydrogen bond cannot be formed by LPyV VP1.
Importance: The most common sialic acid in humans is 5-N-acetyl neuraminic acid (Neu5Ac), but various modifications give rise to more than 50 different sialic acid variants that decorate the cell surface. Unlike most mammals, humans cannot synthesize the sialic acid variant 5-N-glycolyl neuraminic acid (Neu5Gc) due to a gene defect. Humans can, however, still acquire this compound from dietary sources. The role of Neu5Gc in receptor engagement and in defining viral tropism is only beginning to emerge, and structural analyses defining the differences in specificity for Neu5Ac and Neu5Gc are still rare. Using glycan microarray screening and high-resolution protein crystallography, we have examined the receptor specificity of a recently discovered human polyomavirus, HPyV9, and compared it to that of the closely related simian polyomavirus LPyV. Our study highlights critical differences in the specificities of both viruses, contributing to an enhanced understanding of the principles that underlie pathogen selectivity for modified sialic acids.
Figures
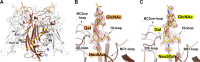


Similar articles
-
Structure analysis of the major capsid proteins of human polyomaviruses 6 and 7 reveals an obstructed sialic acid binding site.J Virol. 2014 Sep;88(18):10831-9. doi: 10.1128/JVI.01084-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008942 Free PMC article.
-
Rules and exceptions: sialic acid variants and their role in determining viral tropism.J Virol. 2014 Jul;88(14):7696-9. doi: 10.1128/JVI.03683-13. Epub 2014 May 7. J Virol. 2014. PMID: 24807712 Free PMC article. Review.
-
Structures of B-lymphotropic polyomavirus VP1 in complex with oligosaccharide ligands.PLoS Pathog. 2013 Oct;9(10):e1003714. doi: 10.1371/journal.ppat.1003714. Epub 2013 Oct 31. PLoS Pathog. 2013. PMID: 24204265 Free PMC article.
-
Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.mBio. 2020 Jul 28;11(4):e00745-20. doi: 10.1128/mBio.00745-20. mBio. 2020. PMID: 32723915 Free PMC article.
-
Production and biomedical applications of virus-like particles derived from polyomaviruses.J Control Release. 2013 Nov 28;172(1):305-321. doi: 10.1016/j.jconrel.2013.08.026. Epub 2013 Aug 31. J Control Release. 2013. PMID: 23999392 Review.
Cited by
-
Structure analysis of the major capsid proteins of human polyomaviruses 6 and 7 reveals an obstructed sialic acid binding site.J Virol. 2014 Sep;88(18):10831-9. doi: 10.1128/JVI.01084-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008942 Free PMC article.
-
Association analyses of large-scale glycan microarray data reveal novel host-specific substructures in influenza A virus binding glycans.Sci Rep. 2015 Oct 28;5:15778. doi: 10.1038/srep15778. Sci Rep. 2015. PMID: 26508590 Free PMC article.
-
Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover.Viruses. 2021 Feb 8;13(2):262. doi: 10.3390/v13020262. Viruses. 2021. PMID: 33567791 Free PMC article. Review.
-
Taking the Scenic Route: Polyomaviruses Utilize Multiple Pathways to Reach the Same Destination.Viruses. 2020 Oct 15;12(10):1168. doi: 10.3390/v12101168. Viruses. 2020. PMID: 33076363 Free PMC article. Review.
-
Rules and exceptions: sialic acid variants and their role in determining viral tropism.J Virol. 2014 Jul;88(14):7696-9. doi: 10.1128/JVI.03683-13. Epub 2014 May 7. J Virol. 2014. PMID: 24807712 Free PMC article. Review.
References
-
- Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257–1260 - PubMed
-
- Gardner SD, Field AM, Coleman DV, Hulme B. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253–1257 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

