Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes
- PMID: 24630722
- PMCID: PMC4000066
- DOI: 10.1016/j.cell.2014.02.008
Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes
Abstract
Inflammasomes elicit host defense inside cells by activating caspase-1 for cytokine maturation and cell death. AIM2 and NLRP3 are representative sensor proteins in two major families of inflammasomes. The adaptor protein ASC bridges the sensor proteins and caspase-1 to form ternary inflammasome complexes, achieved through pyrin domain (PYD) interactions between sensors and ASC and through caspase activation and recruitment domain (CARD) interactions between ASC and caspase-1. We found that PYD and CARD both form filaments. Activated AIM2 and NLRP3 nucleate PYD filaments of ASC, which, in turn, cluster the CARD of ASC. ASC thus nucleates CARD filaments of caspase-1, leading to proximity-induced activation. Endogenous NLRP3 inflammasome is also filamentous. The cryoelectron microscopy structure of ASC(PYD) filament at near-atomic resolution provides a template for homo- and hetero-PYD/PYD associations, as confirmed by structure-guided mutagenesis. We propose that ASC-dependent inflammasomes in both families share a unified assembly mechanism that involves two successive steps of nucleation-induced polymerization. PAPERFLICK:
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
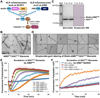
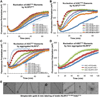
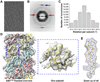

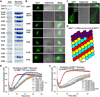


Comment in
-
Inflammasome: putting the pieces together.Cell. 2014 Mar 13;156(6):1127-1129. doi: 10.1016/j.cell.2014.02.038. Cell. 2014. PMID: 24630715
-
Inflammasomes: polymeric assembly.Nat Rev Immunol. 2014 May;14(5):287. doi: 10.1038/nri3669. Epub 2014 Apr 11. Nat Rev Immunol. 2014. PMID: 24722480 No abstract available.
Similar articles
-
The CARD plays a critical role in ASC foci formation and inflammasome signalling.Biochem J. 2013 Feb 1;449(3):613-21. doi: 10.1042/BJ20121198. Biochem J. 2013. PMID: 23110696 Free PMC article.
-
Structural mechanisms of inflammasome assembly.FEBS J. 2015 Feb;282(3):435-44. doi: 10.1111/febs.13133. Epub 2014 Nov 21. FEBS J. 2015. PMID: 25354325 Free PMC article. Review.
-
Self-oligomerization of ASC PYD domain prevents the assembly of inflammasome in vitro.Appl Biochem Biotechnol. 2014 Apr;172(8):3902-12. doi: 10.1007/s12010-014-0819-0. Epub 2014 Mar 2. Appl Biochem Biotechnol. 2014. PMID: 24585381
-
Inhibiting the inflammasome: one domain at a time.Immunol Rev. 2015 May;265(1):205-16. doi: 10.1111/imr.12290. Immunol Rev. 2015. PMID: 25879295 Free PMC article. Review.
-
Purification and analysis of the interactions of caspase-1 and ASC for assembly of the inflammasome.Appl Biochem Biotechnol. 2015 Mar;175(6):2883-94. doi: 10.1007/s12010-014-1471-4. Epub 2015 Jan 8. Appl Biochem Biotechnol. 2015. PMID: 25567507
Cited by
-
Extreme Heat Exposure Induced Acute Kidney Injury through NLRP3 Inflammasome Activation in Mice.Environ Health (Wash). 2024 May 16;2(8):563-571. doi: 10.1021/envhealth.4c00007. eCollection 2024 Aug 16. Environ Health (Wash). 2024. PMID: 39474290 Free PMC article.
-
ASC filament formation serves as a signal amplification mechanism for inflammasomes.Nat Commun. 2016 Jun 22;7:11929. doi: 10.1038/ncomms11929. Nat Commun. 2016. PMID: 27329339 Free PMC article.
-
Contribution of Aberrant Toll Like Receptor Signaling to the Pathogenesis of Myelodysplastic Syndromes.Front Immunol. 2020 Jun 17;11:1236. doi: 10.3389/fimmu.2020.01236. eCollection 2020. Front Immunol. 2020. PMID: 32625214 Free PMC article. Review.
-
As a toxin dies a prion comes to life: A tentative natural history of the [Het-s] prion.Prion. 2015;9(3):184-9. doi: 10.1080/19336896.2015.1038018. Prion. 2015. PMID: 26110610 Free PMC article. Review.
-
Distinct axial and lateral interactions within homologous filaments dictate the signaling specificity and order of the AIM2-ASC inflammasome.Nat Commun. 2021 May 12;12(1):2735. doi: 10.1038/s41467-021-23045-8. Nat Commun. 2021. PMID: 33980849 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

