let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2
- PMID: 24617339
- PMCID: PMC4066225
- DOI: 10.1089/scd.2013.0600
let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2
Abstract
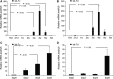
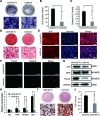
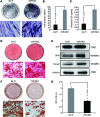
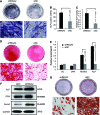
Bone and fat cells share a common progenitor, stromal/mesenchymal stem cells (MSCs), that can differentiate into osteoblasts or adipocytes. Osteogenesis and adipogenesis of MSCs maintain homeostasis under physiological conditions. The disruption of this homeostasis leads to bone-related metabolic diseases. For instance, reduction in bone formation, which is usually accompanied by an increase in bone marrow adipogenesis, occurs with aging, immobility, or osteoporosis. Thus, it is crucial to gain an understanding of how osteogenic and adipogenic lineages of MSCs are regulated. Here, we present evidence that let-7 is a positive regulator of bone development. Using gain- and loss-of-function approaches, we demonstrate that let-7 markedly promotes osteogenesis and suppresses adipogenesis of MSCs in vitro. Moreover, let-7 could promote ectopic bone formation of MSCs in vivo. Subsequent studies further demonstrated that let-7's effects are mediated through the repression of high-mobility group AT-hook 2 (HMGA2) expression. RNAi depletion of HMGA2 could also enhance osteogenesis and repress adipogenesis. Overall, we found a novel role of let-7/HMGA2 axis in regulating the balance of osteogenesis and adipogenesis of MSCs. Thus, let-7 can be used as a novel therapeutic target for disorders that are associated with bone loss and adipocyte accumulation.
Figures


Similar articles
-
Fibroblast Growth Factor 2 Regulates High Mobility Group A2 Expression in Human Bone Marrow-Derived Mesenchymal Stem Cells.J Cell Biochem. 2016 Sep;117(9):2128-37. doi: 10.1002/jcb.25519. Epub 2016 Mar 8. J Cell Biochem. 2016. PMID: 26888666 Free PMC article.
-
MiR-27a is Essential for the Shift from Osteogenic Differentiation to Adipogenic Differentiation of Mesenchymal Stem Cells in Postmenopausal Osteoporosis.Cell Physiol Biochem. 2016;39(1):253-65. doi: 10.1159/000445621. Epub 2016 Jun 24. Cell Physiol Biochem. 2016. PMID: 27337099
-
A novel PPARγ2 modulator sLZIP controls the balance between adipogenesis and osteogenesis during mesenchymal stem cell differentiation.Cell Death Differ. 2014 Oct;21(10):1642-55. doi: 10.1038/cdd.2014.80. Epub 2014 Jun 20. Cell Death Differ. 2014. PMID: 24948012 Free PMC article.
-
The role of microRNAs in cell fate determination of mesenchymal stem cells: balancing adipogenesis and osteogenesis.BMB Rep. 2015 Jun;48(6):319-23. doi: 10.5483/bmbrep.2015.48.6.206. BMB Rep. 2015. PMID: 25341923 Free PMC article. Review.
-
Reciprocal Effect of Environmental Stimuli to Regulate the Adipogenesis and Osteogenesis Fate Decision in Bone Marrow-Derived Mesenchymal Stem Cells (BM-MSCs).Cells. 2023 May 16;12(10):1400. doi: 10.3390/cells12101400. Cells. 2023. PMID: 37408234 Free PMC article. Review.
Cited by
-
miRNA-Based Early Healing Mechanism of Extraction Sockets: miR-190a-5p, a Potential Enhancer of Bone Healing.Biomed Res Int. 2022 Oct 22;2022:7194640. doi: 10.1155/2022/7194640. eCollection 2022. Biomed Res Int. 2022. PMID: 36317115 Free PMC article.
-
Bone marrow mesenchymal stem cell's exosomes as key nanoparticles in osteogenesis and bone regeneration: specific capacity based on cell type.Mol Biol Rep. 2022 Dec;49(12):12203-12218. doi: 10.1007/s11033-022-07807-1. Epub 2022 Oct 12. Mol Biol Rep. 2022. PMID: 36224447 Review.
-
Exosomes: mediators of bone diseases, protection, and therapeutics potential.Oncoscience. 2018 Jun 23;5(5-6):181-195. doi: 10.18632/oncoscience.421. eCollection 2018 May. Oncoscience. 2018. PMID: 30035185 Free PMC article. Review.
-
Cellular and Molecular Connections Between Bone Fracture Healing and Exosomes.Physiol Res. 2023 Nov 28;72(5):565-574. doi: 10.33549/physiolres.935143. Physiol Res. 2023. PMID: 38015756 Free PMC article. Review.
-
FGF8 signaling sustains progenitor status and multipotency of cranial neural crest-derived mesenchymal cells in vivo and in vitro.J Mol Cell Biol. 2015 Oct;7(5):441-54. doi: 10.1093/jmcb/mjv052. Epub 2015 Aug 4. J Mol Cell Biol. 2015. PMID: 26243590 Free PMC article.
References
-
- Fink T, Rasmussen JG, Emmersen J, Pilgaard L, Fahlman A, Brunberg S, Josefsson J, Arnemo JM, Zachar V, Swenson JE. and Frobert O. (2011). Adipose-derived stem cells from the brown bear (Ursus arctos) spontaneously undergo chondrogenic and osteogenic differentiation in vitro. Stem Cell Res 7:89–95 - PubMed
-
- Phinney DG. and Prockop DJ. (2007). Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair—current views. Stem Cells 25:2896–2902 - PubMed
-
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S. and Marshak DR. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147 - PubMed
-
- Prockop DJ. (1997). Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276:71–74 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

