A combinatorial mutagenesis approach for functional epitope mapping on phage-displayed target antigen: application to antibodies against epidermal growth factor
- PMID: 24589624
- PMCID: PMC4011908
- DOI: 10.4161/mabs.28395
A combinatorial mutagenesis approach for functional epitope mapping on phage-displayed target antigen: application to antibodies against epidermal growth factor
Abstract
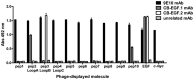
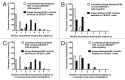
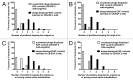
Although multiple different procedures to characterize the epitopes recognized by antibodies have been developed, site-directed mutagenesis remains the method of choice to define the energetic contribution of antigen residues to binding. These studies are useful to identify critical residues and to delineate functional maps of the epitopes. However, they tend to underestimate the roles of residues that are not critical for binding on their own, but contribute to the formation of the target epitope in an additive, or even cooperative, way. Mapping antigenic determinants with a diffuse energetic landscape, which establish multiple individually weak interactions with the antibody paratope, resulting in high affinity and specificity recognition of the epitope as a whole, is thus technically challenging. The current work was aimed at developing a combinatorial strategy to overcome the limitations of site-directed mutagenesis, relying on comprehensive randomization of discrete antigenic regions within phage-displayed antigen libraries. Two model antibodies recognizing epidermal growth factor were used to validate the mapping platform. Abrogation of antibody recognition due to the introduction of simultaneous replacements was able to show the involvement of particular amino acid clusters in epitope formation. The abundance of some of the original residues (or functionally equivalent amino acids sharing their physicochemical properties) among the set of mutated antigen variants selected on a given antibody highlighted their contributions and allowed delineation of a detailed functional map of the corresponding epitope. The use of the combinatorial approach could be expanded to map the interactions between other antigens/antibodies.
Keywords: EGF; library; phage display; randomization; site-directed mutagenesis.
Figures



Similar articles
-
High throughput functional epitope mapping: revisiting phage display platform to scan target antigen surface.MAbs. 2014;6(6):1368-76. doi: 10.4161/mabs.36144. MAbs. 2014. PMID: 25484050 Free PMC article. Review.
-
Epitope mapping by negative selection of randomized antigen libraries displayed on filamentous phage.J Mol Biol. 1997 Jun 27;269(5):704-18. doi: 10.1006/jmbi.1997.1077. J Mol Biol. 1997. PMID: 9223635
-
Epitope mapping: the first step in developing epitope-based vaccines.BioDrugs. 2007;21(3):145-56. doi: 10.2165/00063030-200721030-00002. BioDrugs. 2007. PMID: 17516710 Free PMC article. Review.
-
Deciphering the molecular bases of the biological effects of antibodies against Interleukin-2: a versatile platform for fine epitope mapping.Immunobiology. 2013 Jan;218(1):105-13. doi: 10.1016/j.imbio.2012.02.009. Epub 2012 Feb 16. Immunobiology. 2013. PMID: 22459271
-
Affinity maturation and fine functional mapping of an antibody fragment against a novel neutralizing epitope on human vascular endothelial growth factor.Mol Biosyst. 2013 Aug;9(8):2097-106. doi: 10.1039/c3mb70136k. Epub 2013 May 24. Mol Biosyst. 2013. PMID: 23702826
Cited by
-
Understanding and Modulating Antibody Fine Specificity: Lessons from Combinatorial Biology.Antibodies (Basel). 2022 Jul 14;11(3):48. doi: 10.3390/antib11030048. Antibodies (Basel). 2022. PMID: 35892708 Free PMC article. Review.
-
Deciphering cross-species reactivity of LAMP-1 antibodies using deep mutational epitope mapping and AlphaFold.MAbs. 2023 Jan-Dec;15(1):2175311. doi: 10.1080/19420862.2023.2175311. MAbs. 2023. PMID: 36797224 Free PMC article.
-
Current state of in vivo panning technologies: Designing specificity and affinity into the future of drug targeting.Adv Drug Deliv Rev. 2018 May;130:39-49. doi: 10.1016/j.addr.2018.06.015. Epub 2018 Jun 28. Adv Drug Deliv Rev. 2018. PMID: 29964079 Free PMC article. Review.
-
Molecular and chemical engineering of bacteriophages for potential medical applications.Arch Immunol Ther Exp (Warsz). 2015 Apr;63(2):117-27. doi: 10.1007/s00005-014-0305-y. Epub 2014 Jul 22. Arch Immunol Ther Exp (Warsz). 2015. PMID: 25048831 Free PMC article. Review.
-
Biosensor-based epitope mapping of antibodies targeting the hemagglutinin and neuraminidase of influenza A virus.J Immunol Methods. 2018 Oct;461:23-29. doi: 10.1016/j.jim.2018.07.007. Epub 2018 Jul 24. J Immunol Methods. 2018. PMID: 30053389 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
