The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma
- PMID: 24586173
- PMCID: PMC3937311
- DOI: 10.1371/journal.ppat.1003974
The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma
Abstract
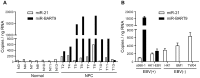
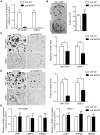
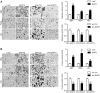
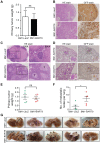
MicroRNAs (miRNAs) are a family of small RNA molecules that negatively regulate the expression of protein-coding genes and play critical roles in orchestrating diverse cellular processes. This regulatory mechanism is also exploited by viruses to direct their life cycle and evade the host immune system. Epstein-Barr virus (EBV) is an oncogenic virus that is closely associated with multiple human diseases, including nasopharyngeal carcinoma (NPC), which is a highly metastatic type of tumor and is frequently reported in South Asia. Several viral proteins have been found to promote the migration and invasiveness of NPC cells. However, not all tumor tissues express these viral oncoproteins, suggesting that other mechanisms may contribute to the aggressive behavior of NPC tumor cells. A previous sequencing study by our group revealed that the EBV miRNA miR-BART9 was expressed at high levels in all EBV-positive NPC tissues. In the present study, we used gain- and loss-of-function approaches to investigate the effect of miR-BART9 in EBV-negative and EBV-positive NPC cells. We discovered that miR-BART9 promotes the migration and invasiveness of cultured NPC cells. The promigratory activity observed in vitro was manifested as an enhanced metastatic ability in vivo. Computational analysis revealed that miR-BART9 may target E-cadherin, a membrane protein that is pivotal in preserving cell-cell junctions and the epithelial phenotype. Through biochemical assays and functional rescue analysis, we confirmed that miR-BART9 specifically inhibits E-cadherin to induce a mesenchymal-like phenotype and promote the migration of NPC cells. These results indicated that miR-BART9 is a prometastatic viral miRNA and suggested that high levels of miR-BART9 in EBV-positive NPC cells may contribute to the aggressiveness of tumor cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
EBV-encoded miRNAs target ATM-mediated response in nasopharyngeal carcinoma.J Pathol. 2018 Apr;244(4):394-407. doi: 10.1002/path.5018. Epub 2018 Feb 16. J Pathol. 2018. PMID: 29230817 Free PMC article.
-
Integrated analysis of the differential cellular and EBV miRNA expression profiles in microdissected nasopharyngeal carcinoma and non-cancerous nasopharyngeal tissues.Oncol Rep. 2015 Nov;34(5):2585-601. doi: 10.3892/or.2015.4237. Epub 2015 Sep 1. Oncol Rep. 2015. PMID: 26330189
-
Epstein-Barr virus BART9 miRNA modulates LMP1 levels and affects growth rate of nasal NK T cell lymphomas.PLoS One. 2011;6(11):e27271. doi: 10.1371/journal.pone.0027271. Epub 2011 Nov 10. PLoS One. 2011. PMID: 22102884 Free PMC article.
-
Emerging roles of small Epstein-Barr virus derived non-coding RNAs in epithelial malignancy.Int J Mol Sci. 2013 Aug 23;14(9):17378-409. doi: 10.3390/ijms140917378. Int J Mol Sci. 2013. PMID: 23979421 Free PMC article. Review.
-
Radio-Susceptibility of Nasopharyngeal Carcinoma: Focus on Epstein- Barr Virus, MicroRNAs, Long Non-Coding RNAs and Circular RNAs.Curr Mol Pharmacol. 2020;13(3):192-205. doi: 10.2174/1874467213666191227104646. Curr Mol Pharmacol. 2020. PMID: 31880267 Review.
Cited by
-
miR-92a-3p Exerts Various Effects in Glioma and Glioma Stem-Like Cells Specifically Targeting CDH1/β-Catenin and Notch-1/Akt Signaling Pathways.Int J Mol Sci. 2016 Oct 27;17(11):1799. doi: 10.3390/ijms17111799. Int J Mol Sci. 2016. PMID: 27801803 Free PMC article.
-
The Function and Therapeutic Potential of Epstein-Barr Virus-Encoded MicroRNAs in Cancer.Mol Ther Nucleic Acids. 2019 Sep 6;17:657-668. doi: 10.1016/j.omtn.2019.07.002. Epub 2019 Jul 15. Mol Ther Nucleic Acids. 2019. PMID: 31400608 Free PMC article. Review.
-
Novel approach to identify putative Epstein-Barr-virus microRNAs regulating host cell genes with relevance in tumor biology and immunology.Oncoimmunology. 2022 May 1;11(1):2070338. doi: 10.1080/2162402X.2022.2070338. eCollection 2022. Oncoimmunology. 2022. PMID: 35529676 Free PMC article.
-
The role of Epstein-Barr virus in epithelial malignancies.J Pathol. 2015 Jan;235(2):323-33. doi: 10.1002/path.4448. J Pathol. 2015. PMID: 25251730 Free PMC article. Review.
-
Epstein-Barr virus-encoded microRNAs involve in tumorigenesis and development.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021 Mar 28;46(3):300-308. doi: 10.11817/j.issn.1672-7347.2021.190744. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 33927078 Free PMC article. Chinese, English.
References
-
- Klein E, Kis LL, Klein G (2007) Epstein-Barr virus infection in humans: from harmless to life endangering virus-lymphocyte interactions. Oncogene 26 (9) 1297–305. - PubMed
-
- Tao Q, Young LS, Woodman CB, Murray PG (2006) Epstein-Barr virus (EBV) and its associated human cancers–genetics, epigenetics, pathobiology and novel therapeutics. Front Biosci 11: 2672–713. - PubMed
-
- Mirzamani N, Salehian P, Farhadi M, Tehran EA (2006) Detection of EBV and HPV in nasopharyngeal carcinoma by in situ hybridization. Exp Mol Pathol 81 (3) 231–4. - PubMed
-
- Tsao SW, Tramoutanis G, Dawson CW, Lo AK, Huang DP (2002) The significance of LMP1 expression in nasopharyngeal carcinoma. Semin Cancer Biol 12 (6) 473–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

