An atlas of chromatoid body components
- PMID: 24554440
- PMCID: PMC3964910
- DOI: 10.1261/rna.043729.113
An atlas of chromatoid body components
Abstract
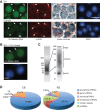
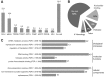
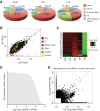
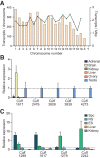
The genome of male germ cells is actively transcribed during spermatogenesis to produce phase-specific protein-coding mRNAs and a considerable amount of different noncoding RNAs. Ribonucleoprotein (RNP) granule-mediated RNA regulation provides a powerful means to secure the quality and correct expression of the requisite transcripts. Haploid spermatids are characterized by a unique, unusually large cytoplasmic granule, the chromatoid body (CB), which emerges during the switch between the meiotic and post-meiotic phases of spermatogenesis. To better understand the role of the CB in male germ cell differentiation, we isolated CBs from mouse testes and revealed its full RNA and protein composition. We showed that the CB is mainly composed of RNA-binding proteins and other proteins involved RNA regulation. The CB was loaded with RNA, including pachytene piRNAs, a diverse set of mRNAs, and a number of uncharacterized long noncoding transcripts. The CB was demonstrated to accumulate nascent RNA during all the steps of round spermatid differentiation. Our results revealed the CB as a large germ cell-specific RNP platform that is involved in the control of the highly complex transcriptome of haploid male germ cells.
Keywords: chromatoid body; male germ cells; piRNA; post-transcriptional; ribonucleoprotein granule.
Figures
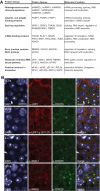
Similar articles
-
Chromatoid body and small RNAs in male germ cells.Reproduction. 2011 Aug;142(2):195-209. doi: 10.1530/REP-11-0057. Epub 2011 Jun 7. Reproduction. 2011. PMID: 21652638 Review.
-
FYCO1 and autophagy control the integrity of the haploid male germ cell-specific RNP granules.Autophagy. 2017 Feb;13(2):302-321. doi: 10.1080/15548627.2016.1261319. Epub 2016 Dec 8. Autophagy. 2017. PMID: 27929729 Free PMC article.
-
The RNA binding protein SAM68 transiently localizes in the chromatoid body of male germ cells and influences expression of select microRNAs.PLoS One. 2012;7(6):e39729. doi: 10.1371/journal.pone.0039729. Epub 2012 Jun 22. PLoS One. 2012. PMID: 22745822 Free PMC article.
-
SMG6 localizes to the chromatoid body and shapes the male germ cell transcriptome to drive spermatogenesis.Nucleic Acids Res. 2022 Nov 11;50(20):11470-11491. doi: 10.1093/nar/gkac900. Nucleic Acids Res. 2022. PMID: 36259644 Free PMC article.
-
Chromatoid Bodies in the Regulation of Spermatogenesis: Novel Role of GRTH.Cells. 2022 Feb 10;11(4):613. doi: 10.3390/cells11040613. Cells. 2022. PMID: 35203264 Free PMC article. Review.
Cited by
-
Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage.BMC Genomics. 2016 Apr 19;17:294. doi: 10.1186/s12864-016-2618-1. BMC Genomics. 2016. PMID: 27094866 Free PMC article.
-
Reproductive Aspects of Chagas Disease Vectors: Evidence of Transcriptional Activity during the Nucleolar Persistence Phenomenon in the Spermatogenesis of Triatomines.Am J Trop Med Hyg. 2019 Sep;101(3):602-604. doi: 10.4269/ajtmh.19-0226. Am J Trop Med Hyg. 2019. PMID: 31359857 Free PMC article.
-
The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis.Nat Commun. 2023 Mar 24;14(1):1636. doi: 10.1038/s41467-023-37252-y. Nat Commun. 2023. PMID: 36964127 Free PMC article.
-
piRNA loading triggers MIWI translocation from the intermitochondrial cement to chromatoid body during mouse spermatogenesis.Nat Commun. 2024 Mar 15;15(1):2343. doi: 10.1038/s41467-024-46664-3. Nat Commun. 2024. PMID: 38491008 Free PMC article.
-
The ReproGenomics Viewer: an integrative cross-species toolbox for the reproductive science community.Nucleic Acids Res. 2015 Jul 1;43(W1):W109-16. doi: 10.1093/nar/gkv345. Epub 2015 Apr 16. Nucleic Acids Res. 2015. PMID: 25883147 Free PMC article.
References
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 - PubMed
-
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods 6: 647–649 - PubMed
-
- Barchi M, Geremia R, Magliozzi R, Bianchi E 2009. Isolation and analyses of enriched populations of male mouse germ cells by sedimentation velocity: The centrifugal elutriation. Methods Mol Biol 558: 299–321 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
