Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside?
- PMID: 24551142
- PMCID: PMC3925152
- DOI: 10.1371/journal.pone.0088716
Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside?
Abstract
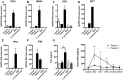
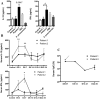
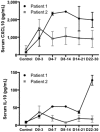
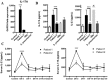
One year after the occurrence of the first case of infection by the Middle East Respiratory Syndrome coronavirus (MERS-CoV) there is no clear consensus on the best treatment to propose. The World Health Organization, as well as several other national agencies, are still working on different clinical approaches to implement the most relevant treatment in MERS-CoV infection. We compared innate and adaptive immune responses of two patients infected with MERS-CoV to understand the underlying mechanisms involved in the response and propose potential therapeutic approaches. Broncho-alveolar lavage (BAL) of the first week and sera of the first month from the two patients were used in this study. Quantitative polymerase chain reaction (qRTPCR) was performed after extraction of RNA from BAL cells of MERS-CoV infected patients and control patients. BAL supernatants and sera were used to assess cytokines and chemokines secretion by enzyme-linked immunosorbent assay. The first patient died rapidly after 3 weeks in the intensive care unit, the second patient still recovers from infection. The patient with a poor outcome (patient 1), compared to patient 2, did not promote type-1 Interferon (IFN), and particularly IFNα, in response to double stranded RNA (dsRNA) from MERS-CoV. The absence of IFNα, known to promote antigen presentation in response to viruses, impairs the development of a robust antiviral adaptive Th-1 immune response. This response is mediated by IL-12 and IFNγ that decreases viral clearance; levels of both of these mediators were decreased in patient 1. Finally, we confirm previous in vitro findings that MERS-CoV can drive IL-17 production in humans. Host recognition of viral dsRNA determines outcome in the early stage of MERS-CoV infection. We highlight the critical role of IFNα in this initial stage to orchestrate a robust immune response and bring substantial arguments for the indication of early IFNα treatment during MERS-CoV infection.
Conflict of interest statement
Figures
Similar articles
-
Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis.J Infect Dis. 2014 May 1;209(9):1331-42. doi: 10.1093/infdis/jit504. Epub 2013 Sep 24. J Infect Dis. 2014. PMID: 24065148 Free PMC article.
-
Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection.mBio. 2019 Mar 26;10(2):e00319-19. doi: 10.1128/mBio.00319-19. mBio. 2019. PMID: 30914508 Free PMC article.
-
MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract.Cytokine. 2020 Feb;126:154895. doi: 10.1016/j.cyto.2019.154895. Epub 2019 Nov 6. Cytokine. 2020. PMID: 31706200 Free PMC article.
-
Modulation of the immune response by Middle East respiratory syndrome coronavirus.J Cell Physiol. 2019 Mar;234(3):2143-2151. doi: 10.1002/jcp.27155. Epub 2018 Aug 26. J Cell Physiol. 2019. PMID: 30146782 Free PMC article. Review.
-
Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Infection, Immunological Response, and Vaccine Development.J Immunol Res. 2019 Apr 7;2019:6491738. doi: 10.1155/2019/6491738. eCollection 2019. J Immunol Res. 2019. PMID: 31089478 Free PMC article. Review.
Cited by
-
The Interplay between Airway Cilia and Coronavirus Infection, Implications for Prevention and Control of Airway Viral Infections.Cells. 2024 Aug 14;13(16):1353. doi: 10.3390/cells13161353. Cells. 2024. PMID: 39195243 Free PMC article. Review.
-
T-Cell Epitope Mapping of SARS-CoV-2 Reveals Coordinated IFN-γ Production and Clonal Expansion of T Cells Facilitates Recovery from COVID-19.Viruses. 2024 Jun 22;16(7):1006. doi: 10.3390/v16071006. Viruses. 2024. PMID: 39066169 Free PMC article.
-
Characterization and immunogenicity assessment of MERS-CoV pre-fusion spike trimeric oligomers as vaccine immunogen.Hum Vaccin Immunother. 2024 Dec 31;20(1):2351664. doi: 10.1080/21645515.2024.2351664. Epub 2024 May 17. Hum Vaccin Immunother. 2024. PMID: 38757508 Free PMC article.
-
Plasma of COVID-19 Patients Does Not Alter Electrical Resistance of Human Endothelial Blood-Brain Barrier In Vitro.Function (Oxf). 2024 Jan 9;5(2):zqae002. doi: 10.1093/function/zqae002. eCollection 2024. Function (Oxf). 2024. PMID: 38486975 Free PMC article.
-
Enhanced antiviral immunity and dampened inflammation in llama lymph nodes upon MERS-CoV sensing: bridging innate and adaptive cellular immune responses in camelid reservoirs.Front Immunol. 2023 Jun 14;14:1205080. doi: 10.3389/fimmu.2023.1205080. eCollection 2023. Front Immunol. 2023. PMID: 37388723 Free PMC article.
References
-
- Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, et al. (2013) Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13: 752–761 Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id.... - PMC - PubMed
-
- Katze MG, He Y, Gale M (2002) Viruses and interferon: a fight for supremacy. Nature Reviews Immunology 2: 675–687 Available: http://www.nature.com/doifinder/10.1038/nri888. - DOI - PubMed
-
- Takeuchi O, Akira S (2008) MDA5/RIG-I and virus recognition. Current opinion in immunology 20: 17–22 Available: http://linkinghub.elsevier.com/retrieve/pii/S0952791508000046. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

