Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation
- PMID: 24532714
- PMCID: PMC3937514
- DOI: 10.1101/gad.231407.113
Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation
Abstract
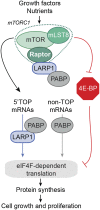
The mammalian target of rapamycin (mTOR) promotes cell growth and proliferation by promoting mRNA translation and increasing the protein synthetic capacity of the cell. Although mTOR globally promotes translation by regulating the mRNA 5' cap-binding protein eIF4E (eukaryotic initiation factor 4E), it also preferentially regulates the translation of certain classes of mRNA via unclear mechanisms. To help fill this gap in knowledge, we performed a quantitative proteomic screen to identify proteins that associate with the mRNA 5' cap in an mTOR-dependent manner. Using this approach, we identified many potential regulatory factors, including the putative RNA-binding protein LARP1 (La-related protein 1). Our results indicate that LARP1 associates with actively translating ribosomes via PABP and that LARP1 stimulates the translation of mRNAs containing a 5' terminal oligopyrimidine (TOP) motif, encoding for components of the translational machinery. We found that LARP1 associates with the mTOR complex 1 (mTORC1) and is required for global protein synthesis as well as cell growth and proliferation. Together, these data reveal important molecular mechanisms involved in TOP mRNA translation and implicate LARP1 as an important regulator of cell growth and proliferation.
Keywords: 5′TOP; LARP1; mRNA; mTOR; proteomics; translation.
Figures





Similar articles
-
La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1).J Biol Chem. 2015 Jun 26;290(26):15996-6020. doi: 10.1074/jbc.M114.621730. Epub 2015 May 4. J Biol Chem. 2015. PMID: 25940091 Free PMC article.
-
La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs.Elife. 2017 Apr 7;6:e24146. doi: 10.7554/eLife.24146. Elife. 2017. PMID: 28379136 Free PMC article.
-
Distinct roles of LARP1 and 4EBP1/2 in regulating translation and stability of 5'TOP mRNAs.Sci Adv. 2024 Feb 16;10(7):eadi7830. doi: 10.1126/sciadv.adi7830. Epub 2024 Feb 16. Sci Adv. 2024. PMID: 38363833 Free PMC article.
-
LARP1 and LARP4: up close with PABP for mRNA 3' poly(A) protection and stabilization.RNA Biol. 2021 Feb;18(2):259-274. doi: 10.1080/15476286.2020.1868753. Epub 2021 Jan 31. RNA Biol. 2021. PMID: 33522422 Free PMC article. Review.
-
Controversies around the function of LARP1.RNA Biol. 2021 Feb;18(2):207-217. doi: 10.1080/15476286.2020.1733787. Epub 2020 Apr 1. RNA Biol. 2021. PMID: 32233986 Free PMC article. Review.
Cited by
-
Targeting the eIF4F translation initiation complex: a critical nexus for cancer development.Cancer Res. 2015 Jan 15;75(2):250-63. doi: 10.1158/0008-5472.CAN-14-2789. Cancer Res. 2015. PMID: 25593033 Free PMC article. Review.
-
Decoding ribosome complexity: role of ribosomal proteins in cancer and disease.NAR Cancer. 2024 Jul 23;6(3):zcae032. doi: 10.1093/narcan/zcae032. eCollection 2024 Sep. NAR Cancer. 2024. PMID: 39045153 Free PMC article.
-
Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome.Nat Cell Biol. 2022 Mar;24(3):299-306. doi: 10.1038/s41556-022-00852-9. Epub 2022 Mar 15. Nat Cell Biol. 2022. PMID: 35292784 Free PMC article.
-
Sorting mRNA Molecules for Cytoplasmic Transport and Localization.Front Genet. 2018 Nov 6;9:510. doi: 10.3389/fgene.2018.00510. eCollection 2018. Front Genet. 2018. PMID: 30459808 Free PMC article. Review.
-
Surviving and Adapting to Stress: Translational Control and the Integrated Stress Response.Antioxid Redox Signal. 2023 Aug;39(4-6):351-373. doi: 10.1089/ars.2022.0123. Epub 2023 May 9. Antioxid Redox Signal. 2023. PMID: 36943285 Free PMC article. Review.
References
-
- Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al. 2012. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res 72: 6468–6476 - PubMed
-
- Aoki K, Adachi S, Homoto M, Kusano H, Koike K, Natsume T 2013. LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett 587: 2173–2178 - PubMed
-
- Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM 2009. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol 334: 186–197 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
