Dedifferentiation and reprogramming: origins of cancer stem cells
- PMID: 24531722
- PMCID: PMC3989690
- DOI: 10.1002/embr.201338254
Dedifferentiation and reprogramming: origins of cancer stem cells
Abstract
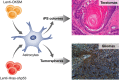
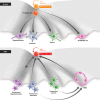
Regenerative medicine aims to replace the lost or damaged cells in the human body through a new source of healthy transplanted cells or by endogenous repair. Although human embryonic stem cells were first thought to be the ideal source for cell therapy and tissue repair in humans, the discovery by Yamanaka and colleagues revolutionized the field. Almost any differentiated cell can be sent back in time to a pluripotency state by expressing the appropriate transcription factors. The process of somatic reprogramming using Yamanaka factors, many of which are oncogenes, offers a glimpse into how cancer stem cells may originate. In this review we discuss the similarities between tumor dedifferentiation and somatic cell reprogramming and how this may pose a risk to the application of this new technology in regenerative medicine.
Figures

Similar articles
-
Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration.Nat Rev Mol Cell Biol. 2011 Feb;12(2):79-89. doi: 10.1038/nrm3043. Nat Rev Mol Cell Biol. 2011. PMID: 21252997 Review.
-
The dualistic origin of human tumors.Semin Cancer Biol. 2018 Dec;53:1-16. doi: 10.1016/j.semcancer.2018.07.004. Epub 2018 Jul 21. Semin Cancer Biol. 2018. PMID: 30040989 Free PMC article. Review.
-
Induced pluripotency: history, mechanisms, and applications.Genes Dev. 2010 Oct 15;24(20):2239-63. doi: 10.1101/gad.1963910. Genes Dev. 2010. PMID: 20952534 Free PMC article. Review.
-
Reprogramming of retinoblastoma cancer cells into cancer stem cells.Biochem Biophys Res Commun. 2017 Jan 22;482(4):549-555. doi: 10.1016/j.bbrc.2016.11.072. Epub 2016 Nov 14. Biochem Biophys Res Commun. 2017. PMID: 27856246
-
Transdifferentiation and reprogramming: Overview of the processes, their similarities and differences.Biochim Biophys Acta Mol Cell Res. 2017 Jul;1864(7):1359-1369. doi: 10.1016/j.bbamcr.2017.04.017. Epub 2017 Apr 28. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 28460880 Review.
Cited by
-
Epigenetic priming of an epithelial enhancer by p63 and CTCF controls expression of a skin-restricted gene XP33.Cell Death Discov. 2023 Dec 8;9(1):446. doi: 10.1038/s41420-023-01716-3. Cell Death Discov. 2023. PMID: 38065940 Free PMC article.
-
A Greedy Algorithm-Based Stem Cell LncRNA Signature Identifies a Novel Subgroup of Lung Adenocarcinoma Patients With Poor Prognosis.Front Oncol. 2020 Aug 11;10:1203. doi: 10.3389/fonc.2020.01203. eCollection 2020. Front Oncol. 2020. PMID: 32850350 Free PMC article.
-
The Transcriptomic Landscape of Mismatch Repair-Deficient Intestinal Stem Cells.Cancer Res. 2021 May 15;81(10):2760-2773. doi: 10.1158/0008-5472.CAN-20-2896. Epub 2021 Mar 18. Cancer Res. 2021. PMID: 34003775 Free PMC article.
-
Reprogramming strategies for the establishment of novel human cancer models.Cell Cycle. 2016 Sep 16;15(18):2393-7. doi: 10.1080/15384101.2016.1196305. Epub 2016 Jun 17. Cell Cycle. 2016. PMID: 27314153 Free PMC article. Review.
-
MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat.Nat Commun. 2019 Aug 9;10(1):3623. doi: 10.1038/s41467-019-11568-0. Nat Commun. 2019. PMID: 31399583 Free PMC article.
References
-
- Kaiser LR. The future of multihospital systems. Top Health Care Financ. 1992;18:32–45. - PubMed
-
- Lysaght MJ, Crager J. Origins. Tissue Eng Part A. 2009;15:1449–1450. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

